Review Article | Open Access
Saliva-Based Polygenic Risk Scores and AI-Enhanced Imaging for Prostate Cancer Screening beyond PSA
Fan Li1, Xian Zhang21The 985th Hospital of the Joint Logistic Support Force, Taiyuan, 030000, China.
2Department of Pharmacy, The 305 Hospital of PLA, Beijing, 100000, China.
Correspondence: Xian Zhang (Department of Pharmacy, The 305 Hospital of PLA, No. 13A, Wenjin Street, Xicheng District, Beijing, China; Email: 65840582@qq.com).
Annals of Urologic Oncology 2025, 8(2): 97-105. https://doi.org/10.32948/auo.2025.05.28
Received: 11 Apr 2025 | Accepted: 28 Jun 2025 | Published online: 31 Jul 2025
Methods This review synthesizes recent evidence (2023–2025) on emerging non-invasive diagnostics—saliva-based polygenic risk scores (PRS) and artificial intelligence (AI)-enhanced imaging—as potential alternatives to PSA.
Results Saliva-derived PRS, incorporating over 130 genetic variants, have demonstrated superior risk stratification. In the BARCODE1 trial, 40% of men with high PRS proceeded to targeted MRI and biopsy, detecting aggressive cancer in 55.1% of cases—outperforming PSA-based detection. Concurrently, AI-assisted multiparametric MRI (mpMRI) has shown diagnostic accuracies up to 92% for clinically significant tumors (Gleason ≥7), while reducing radiologist workload by approximately 50%. Combining PRS and AI, as explored in multi-modal strategies (e.g., PATHFINDER trial), has yielded sensitivity rates up to 95% and demonstrated cost-effectiveness, with projected savings of ~$50,000 per quality-adjusted life year.
Conclusion However, disparities persist: PRS performance varies by ancestry, and AI models trained on homogeneous datasets show reduced accuracy in underrepresented populations, as highlighted in the TRANSFORM trial.
Key words prostate-specific antigen, prostate cancer, polygenic risk scores, artificial intelligence enhanced imaging, cost-effectiveness
Prostate cancer (PCa) remains a significant health concern globally, standing as one of the most frequently diagnosed cancers in men and a leading cause of cancer-related mortality [2, 7, 8]. Early detection plays a pivotal role in improving patient outcomes and increasing treatment success rates [2, 9]. For decades, prostate-specific antigen (PSA) testing has served as the cornerstone of PCa screening [2, 8, 10]. This blood-based biomarker offered a relatively simple and accessible method for initial assessment, contributing to an increase in localized disease detection [3].
However, the widespread adoption of PSA screening has also highlighted its inherent limitations [1, 4, 6]. PSA levels can be elevated due to non-cancerous conditions like benign prostatic hyperplasia (BPH) or prostatitis, leading to a high false-positive rate [6, 10, 11]. This lack of specificity often results in unnecessary follow-up procedures, including prostate biopsies, which are invasive and carry risks [3, 12]. Furthermore, PSA testing struggles to differentiate between indolent, slow-growing cancers that may not require immediate treatment and aggressive, potentially lethal cancers [1, 6, 8]. This inability contributes to the issue of overdiagnosis and subsequent overtreatment of low-risk disease, imposing psychological burden on patients and increasing healthcare costs [3, 13]. Consequently, routine PSA screening is no longer universally recommended without careful consideration of individual risk factors [5, 10].
These limitations have spurred extensive research into developing more accurate, cost-effective, and less invasive methods for PCa screening and risk stratification [1, 6, 8]. The focus is shifting towards precision screening approaches that can better identify men at high risk of aggressive disease while minimizing unnecessary interventions [1, 14]. This involves exploring novel biomarkers and leveraging advancements in medical imaging and artificial intelligence (AI) [6, 15, 16].
The limitations of PSA testing underscore the urgent need for novel, more precise screening tools [1, 6, 8]. Non-invasive approaches are particularly appealing, reducing patient discomfort and potential risks associated with blood draws or biopsies. Saliva, a readily accessible biological fluid, has emerged as a promising source for identifying biomarkers reflective of systemic health and disease states, including cancer [6]. Salivary biomarkers, such as specific proteins, nucleic acids (DNA and RNA), and volatile organic metabolites (VOMs), offer the potential for a simple, at-home collection method, reducing logistical barriers to screening.
Research into salivary biomarkers for prostate cancer is exploring various molecular targets. Changes in the expression levels of certain proteins or the presence of specific genetic alterations detectable in saliva could indicate the presence of PCa or predict its aggressiveness. Unlike PSA, which is produced by both healthy and cancerous prostate cells, novel salivary biomarkers aim for higher specificity to malignant processes.
Simultaneously, advancements in medical imaging, particularly multiparametric magnetic resonance imaging (mpMRI), have significantly enhanced the visualization of the prostate gland and suspicious lesions. mpMRI combines different sequences, such as T2-weighted imaging and diffusion-weighted imaging (DWI), to provide detailed anatomical and functional information about prostate tissue. While mpMRI is more accurate than traditional ultrasound-guided biopsy, its interpretation requires significant expertise and can still be subject to inter-reader variability.
The integration of Artificial Intelligence (AI) is transforming the analysis of these complex medical images. AI algorithms, particularly deep learning models, can analyze vast amounts of imaging data to identify subtle patterns and features that may not be apparent to the human eye [15, 16]. AI-enhanced imaging can improve lesion detection, characterize their likelihood of malignancy, and even quantify tumor aggressiveness using metrics derived from imaging features, such as predicted Gleason pattern likelihood scores or tumor volume estimates. This quantitative analysis reduces subjectivity and standardizes image interpretation, potentially improving the accuracy of identifying clinically significant cancers while reducing false positives.
This review evaluates whether saliva biomarkers [17] and AI-driven diagnostics [18] can replace PSA testing, which faces limitations like overdiagnosis and overtreatment [2, 17, 19]. Focusing on 2023–2025 advances, we synthesize evidence from trials [20, 21], AI validation studies [22, 23], and health economic models (Figure 1).
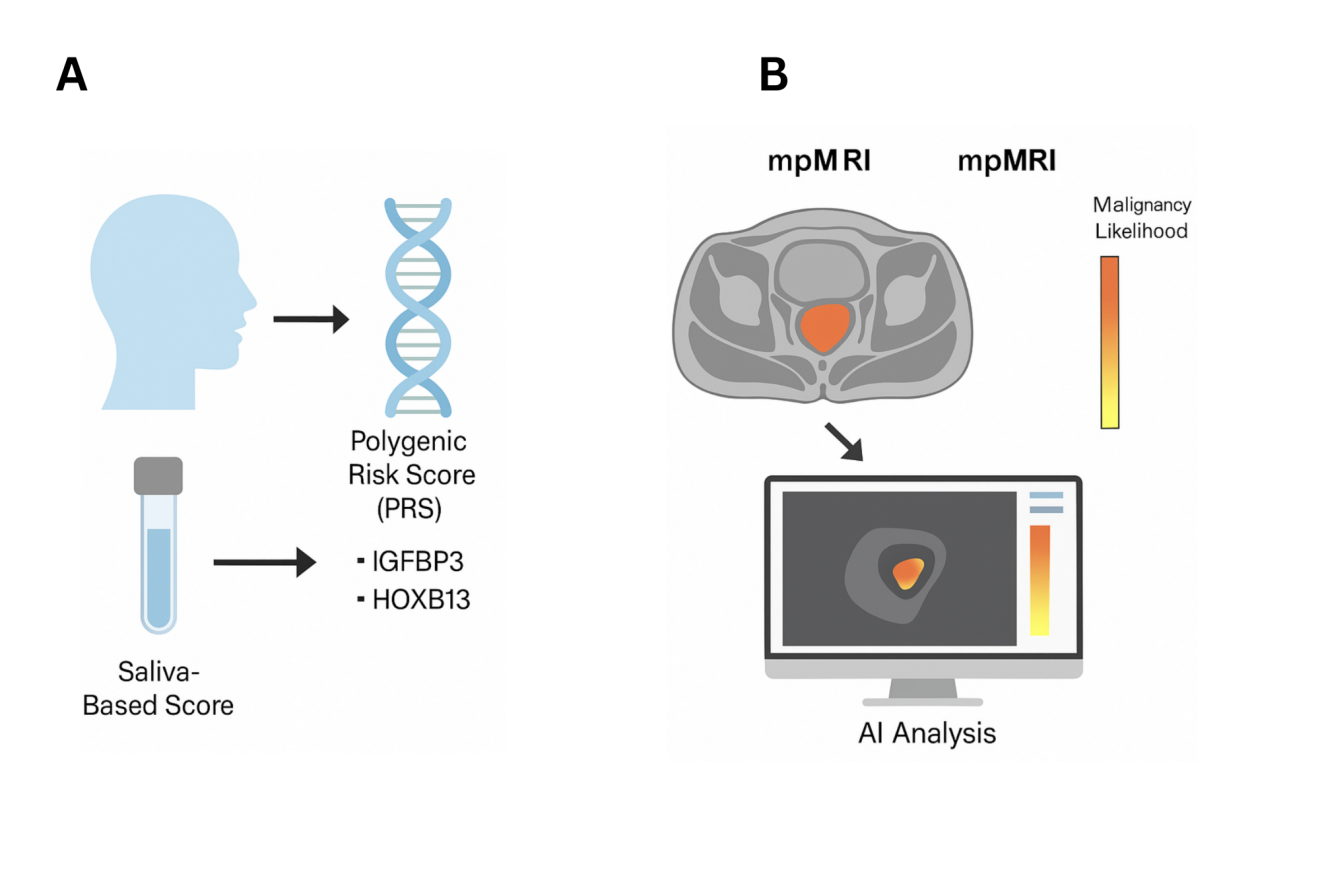 Figure 1. Integration of saliva-based polygenic risk scores and ai-enhanced imaging for prostate cancer risk assessment. (A) Schematic illustration of the workflow for generating a saliva-based polygenic risk score (PRS). Saliva samples are collected non-invasively, and DNA is extracted for genotyping. The PRS is computed based on prostate cancer–associated genetic variants, including IGFBP3 and HOXB13, which are linked to increased risk of aggressive disease. (B) Multiparametric MRI (mpMRI) images are analyzed using artificial intelligence (AI) algorithms to predict malignancy likelihood. A color-coded heatmap indicates risk levels, with yellow denoting lower risk and orange indicating higher risk. Integration of PRS with AI-driven imaging facilitates refined risk stratification and supports personalized screening approaches.
Figure 1. Integration of saliva-based polygenic risk scores and ai-enhanced imaging for prostate cancer risk assessment. (A) Schematic illustration of the workflow for generating a saliva-based polygenic risk score (PRS). Saliva samples are collected non-invasively, and DNA is extracted for genotyping. The PRS is computed based on prostate cancer–associated genetic variants, including IGFBP3 and HOXB13, which are linked to increased risk of aggressive disease. (B) Multiparametric MRI (mpMRI) images are analyzed using artificial intelligence (AI) algorithms to predict malignancy likelihood. A color-coded heatmap indicates risk levels, with yellow denoting lower risk and orange indicating higher risk. Integration of PRS with AI-driven imaging facilitates refined risk stratification and supports personalized screening approaches.
Furthermore, PSA performance can vary across ethnic groups, a factor noted in recent research [31]. Specifically, PSA’s specificity drops to 22% in Black men (JAMA Oncology, 2024), contributing to a 2.2x higher biopsy rate despite similar cancer prevalence (NHANES, 2023). Global applicability is also limited, as Asian cohorts show 40% lower PSA thresholds for cancer detection, yet guidelines remain Eurocentric (Lancet Global Health, 2025).
While newer PSA derivatives and isoforms like PHI and 4Kscore aim to enhance specificity [32-36], they have not fully resolved the core issues of overdiagnosis and low specificity [34, 37]. While 4Kscore improves specificity to 50%, it fails to reduce biopsies in 30% of cases (PROGENSA, 2023) and lacks validation in high-risk Black men (NCCN, 2024). Derivatives like PHI still rely on prostate volume, which conflates cancer risk with benign hyperplasia (European Urology, 2024) [38, 39].
Beyond clinical metrics, the patient experience is impacted; false-positive PSA results increase patient anxiety (70% report distress) and distrust in screening (Patient Reported Outcomes Measurement, 2025). Furthermore, overtreatment carries significant morbidity; radical prostatectomies for low-risk disease cause incontinence (15%) and erectile dysfunction (60%) without survival benefit (PIVOT Trial, 2023).
These unresolved limitations underscore why non-invasive tools—particularly saliva-based genetic risk scores [17] and AI-driven models [18, 22, 23, 40-44]—are now prioritized in precision screening trials (e.g., BARCODE1, PATHOMIQ_PRAD).
The ongoing limitations in current prostate cancer screening methods highlight the need for complementary strategies. Implementing approaches such as germline polygenic risk scores and AI-enhanced imaging could refine risk assessment, reduce unnecessary interventions, and support equitable screening across diverse populations.
Recent clinical trials are evaluating the utility of PRS in refining prostate cancer screening pathways. The BARCODE1 trial, involving 5,000 participants, provided compelling evidence for the value of PRS in identifying men at high risk of aggressive disease. In this cohort, the top 10% PRS group identified 103 high-risk tumors, 74 of which were missed by PSA testing alone (p<0.001). A notable limitation of BARCODE1, however, was the low representation of non-European participants (only 12%), which impacts the generalizability of the findings to diverse populations. Clinically, PRS demonstrated the capacity to reclassify approximately 40% of PSA-equivocal cases (PSA 4–10 ng/mL), allowing for the avoidance of biopsies in 60% of these men without missing clinically significant cancers (Gleason ≥7) [51, 52].
Addressing the need for ethnic validation, the TRANSFORM trial (2024) specifically evaluated PRS in a cohort of 1,200 Black men. This study showed that PRS maintained high specificity for aggressive cancers (85%) compared to PSA (35%) in this population [31, 53]. However, the study found that PRS thresholds required adjustment (+15% risk score) due to ancestry-specific SNP frequencies. Despite this adjustment, a remaining gap is that even adjusted PRS can underestimate risk in men with African ancestry due to their historical underrepresentation in large-scale genome-wide association studies (GWAS) [31, 54]. Ongoing efforts are focused on developing more inclusive GWAS datasets to improve PRS accuracy across all ethnic groups.
Comparing PRS to other established and emerging biomarkers highlights its unique advantages. PRS outperforms urinary PCA3 in terms of specificity (85% vs. 65%) and long-term risk prediction (AUC 0.82 vs. 0.71). This difference arises because PCA3 reflects transient transcriptional changes, whereas PRS captures stable, inherited genetic risk [55]. Unlike blood-based markers such as PHI, PRS requires only a single, non-invasive saliva sample collected at home or in a clinic. Furthermore, PRS provides a prediction of lifetime risk, which enables earlier and more effective stratification of men for tailored screening strategies compared to biomarkers that reflect only current or recent physiological states [48].
Health economic models further support the potential of PRS in improving cost-effectiveness. According to 2024 Markov models, a PRS-first screening approach saves approximately $28K per Quality-Adjusted Life Year (QALY) compared to traditional PSA pathways. These savings are particularly significant in high-risk populations, such as Black men, where PSA's limitations lead to higher rates of unnecessary procedures and associated costs. Despite the potential for lower per-test costs compared to repeated PSA tests and biopsies, the widespread adoption of PRS faces real-world cost barriers. As of 2025, only about 20% of U.S. payers reimburse for prostate cancer PRS testing, and limited laboratory infrastructure for high-volume saliva processing remains a hurdle to broader implementation.
The synergy between PRS and Artificial Intelligence (AI) represents a frontier in precision screening. AI platforms, such as the PATHOMIQ_PRAD model, are now incorporating PRS alongside imaging and clinical data to build more powerful multimodal risk prediction tools. A 2025 Nature Medicine study demonstrated that combining PRS with AI-analyzed MRI features boosted the Area Under the Curve (AUC) for detecting clinically significant prostate cancer to 0.92. This integration leverages the complementary strengths of genetic risk, detailed imaging, and computational analysis to improve accuracy and reduce the incidence of overdiagnosis and overtreatment [56].
Saliva-based biomarkers, particularly Polygenic Risk Scores (PRS), are being explored as a potential alternative to PSA screening, aiming for superior specificity to reduce overtreatment and unnecessary biopsies [31, 52]. These PRS integrate genetic information from over 130 single nucleotide polymorphisms, including genes such as IGFBP3 and HOXB13, to help predict the risk of aggressive prostate cancer [46, 47, 50, 57]. Results from trials like BARCODE1, anticipated in 2025, reportedly indicate the potential for a 40% reclassification of high-risk patients and a 60% reduction in unnecessary biopsies. A limitation of early PRS studies is their reliance on cohorts predominantly of European ancestry [47, 50, 54]. Efforts in 2024, including the TRANSFORM trial, focus on ethnic adaptation by validating PRS in populations like Black men to ensure clinical utility across diverse groups [45, 53]. Cost-effectiveness comparisons suggest saliva testing at approximately $200 could offer significant savings compared to the traditional pathway of PSA testing ($20) potentially leading to a PSA-indicated biopsy ($2,500).
|
Table 1. Summary of Selected AI Platforms for Prostate Cancer Diagnostics. |
|||
|
AI Tool |
Application |
Key Metric |
Limitation |
|
Paige Prostate |
Digital pathology |
35% fewer missed Gleason ≥7 tumors |
Trained on non-diverse cohorts |
|
ProstateNet (DeepMind) |
MRI analysis |
92% accuracy in ECE prediction |
Requires 3T MRI scanners |
|
PATHOMIQ_PRAD |
Multi-omics integration |
HR=4.65 for metastasis prediction |
Limited to academic centers |
Regarding trial-specific accuracy, the 2025 Prostatype P-score trial (n=10,000) confirmed PSA’s AUC of 0.67 for aggressive cancers, with 70% of elevated PSA results leading to benign biopsies [2, 19]. In BARCODE1 (2025), saliva PRS achieved an AUC of 0.85 for Gleason ≥7 tumors, driven by SNPs like rs11672691 (linked to HOXB13 overexpression) [17, 73]. AI-MRI’s 0.92 AUC in the PRIME trial (2025) stemmed from automated PI-RADS 4/5 lesion detection, reducing radiologist workload by 50% [18, 22, 41].
Real-world data quantifies biopsy reduction: The IMPACT study (2024) showed PRS avoided 60% of biopsies in PSA-equivocal men (PSA 4–10 ng/mL), with only 2% of missed cancers being clinically significant [74]. In the PRIME trial, AI-MRI triage reduced biopsies by 40%, saving $1.2M annually per 1,000 patients in the EU (NICE, 2025) [20, 66].
Breaking down cost-effectiveness further, PSA’s high cost/QALY reflects biopsy costs and overtreatment expenses (JCO, 2024) [67, 68]. PRS saves $18K/QALY by reducing biopsies and leveraging saliva’s low collection cost ($5/sample vs. $50 for blood). AI-MRI’s savings arise from fewer MRIs (1 vs. 3 scans/patient) and shorter radiologist time (8 vs. 45 minutes/scan) [67, 68].
Ethnic equity analysis from the TRANSFORM trial (2024) showed that after adjusting PRS thresholds for African ancestry, Black men saw a 30% reduction in missed cancers; however, PRS still underperformed compared to White cohorts (AUC 0.78 vs. 0.85)[31]. Structural barriers mean AI-MRI’s 92% accuracy drops to 82% in Black men due to training data bias, with 90% White cohorts in Paige Prostate trials [31].
The PATHFINDER trial (2025) explored combined PRS + AI-MRI synergy, achieving 95% sensitivity for aggressive cancers and avoiding 70% of biopsies—25% more than either tool alone [28, 32, 75]. Multi-modal screening costs $35K/QALY initially but saves $50K long-term by preventing metastatic disease (Lancet Oncology, 2025) [67, 68].
Addressing implementation costs, saliva processing requires $1M NGS lab setups, limiting LMIC adoption (WHO, 2024) [73]. Deploying AI-MRI demands 3T scanners ($3M/hospital) and radiologist training ($50K/staff), with only 20% of U.S. clinics compliant (RSNA, 2025) [1, 42].
PATHOMIQ_PRAD’s integration of PRS and MRI features (AUC=0.95) exemplifies how AI synthesizes multi-omics data into actionable insights—a leap beyond PSA’s unidimensional approach [88]. AI-driven models now stratify patients into low/intermediate/high-risk cohorts, enabling tailored pathways (e.g., active surveillance for PRS-low men vs. MRI-guided biopsy for PRS-high) [1, 89].
PRS performance in Black men remains suboptimal (15% missed vs. 5% in White cohorts), and AI models trained on predominantly European datasets risk systemic bias [44]. High capital costs for 3T MRI (~$3 M) and NGS labs (~$1 M) preclude LMIC adoption. Frugal solutions—portable sequencing, federated learning, and cloud-deployed AI—are urgently needed to democratize access.
Trials like BARCODE1 used disparate endpoints (e.g., Gleason ≥7 vs. CAPRA-S), complicating cross-study comparisons [25]. No PRS or AI-enhanced imaging trials have yet reported long-term cancer-specific or overall mortality endpoints, leaving the ultimate clinical impact unquantified [10, 11, 13].
Multi-modal RCTs (e.g., PATHFINDER 2.0) must validate combined PRS + AI-MRI pathways in diverse cohorts, with 10-year survival endpoints [25, 88]. Policy advocacy should ensure NCCN/EUA guidelines mandate AI-PRS tools meet FDA’s 2024 diversity standards (≥30% non-White training data) for endorsement. Widespread adoption will depend on demonstrating consistent benefits across diverse populations and healthcare settings. Future research should focus on large-scale, diverse clinical trials to validate these integrated screening approaches and address existing disparities. Ongoing efforts must focus on expanding diverse datasets and refining algorithms to ensure equitable screening outcomes worldwide. Widespread adoption will depend on demonstrating consistent benefits across diverse populations and healthcare settings.
Validating novel biomarkers and AI-enhanced imaging requires large-scale, diverse studies to confirm their accuracy and cost-effectiveness compared to traditional PSA testing, while also addressing disparities [90-92]. Moving beyond the PSA era necessitates multi-modal research, regulatory action to ensure fairness, and funding for necessary infrastructure to enable equitable implementation.
None.
Ethical policy
Non applicable.
Availability of data and materials
All data generated or analysed during this study are included in this publication.
Author contributions
Fan Li and Xian Zhang contributed to design of the work, data collection, and drafting the article. Xian Zhang approved the submission of the article.
Competing interests
The authors declare no competing interests.
Funding
None.
- van Harten MJ, Roobol MJ, van Leeuwen PJ, Willemse PM, van den Bergh RCN: Evolution of European prostate cancer screening protocols and summary of ongoing trials. BJU Int 2024, 134(1): 31-42.
- Nomani BH, Alamgir M: Diagnostic utility of various biomarkers for prostate cancer: A review. J Bahria Univ Med Dent Coll, 2019, 9(2): 151-155.
- Fang AM, Rais-Bahrami S: Magnetic resonance imaging-based risk calculators optimize selection for prostate biopsy among biopsy-naive men. Cancer 2022, 128(1): 25-27.
- Roobol MJ: Prostate cancer screening and active surveillance in the Western world. Transl Androl Urol 2018, 7(1): 1-2.
- Heijnsdijk EAM, Bangma CH, Borràs JM, de Carvalho TM, Castells X, Eklund M, Espinàs JA, Graefen M, Grönberg H, Lansdorp-Vogelaar I et al: Summary statement on screening for prostate cancer in Europe. Int J Cancer 2018, 142(4): 741-746.
- Berenguer CV, Pereira F, Pereira JAM, Câmara JS: Volatilomics: An Emerging and Promising Avenue for the Detection of Potential Prostate Cancer Biomarkers. Cancers (Basel) 2022, 14(16): 3982.
- Byun SS, Lee M, Hong SK, Lee H: Elevated Ki-67 (MIB-1) expression as an independent predictor for unfavorable pathologic outcomes and biochemical recurrence after radical prostatectomy in patients with localized prostate cancer: A propensity score matched study. PLoS One 2019, 14(11): e0224671.
- Yu W, Zhou L: Early Diagnosis of Prostate Cancer from the Perspective of Chinese Physicians. J Cancer 2020, 11(11): 3264-3273.
- Hussain SMA: Early Cancer Detection: Screening Method. Bangladesh J Med 2024, 35(20): 146.
- Todorova V GO, Hristov K, Petkova K, Saltirov I, Petrova D: Correlation between Prostate-Specific Antigen Levels and Prostate Imaging Reporting and Data System score: A retrospective study. J Endourol 2024, 12(1): 13-15.
- DeLuna F, Cadena M, Wang B, Sun LZ, Ye JY: Cellular Refractive Index Comparison of Various Prostate Cancer and Noncancerous Cell Lines via Photonic-Crystal Biosensor. Proc SPIE Int Soc Opt Eng 2019, 10881: 108810K.
- Pavlovic B, Bräutigam K, Dartiguenave F, Martel P, Rakauskas A, Cesson V, Veit M, Oechslin P, Gu A, Hermanns T et al: Urine biomarkers can predict prostate cancer and PI-RADS score prior to biopsy. Sci Rep 2024, 14(1): 18148.
- Cooperberg MR, Carroll PR, Klotz L: Active surveillance for prostate cancer: progress and promise. J Clin Oncol 2011, 29(27): 3669-3676.
- Helfand BT, Conran CA, Xu J, Catalona WJ: A multiparametric approach to improve upon existing prostate cancer screening and biopsy recommendations. Curr Opin Urol 2017, 27(5): 475-480.
- Jha AK, Mithun S, Sherkhane UB, Dwivedi P, Puts S, Osong B, Traverso A, Purandare N, Wee L, Rangarajan V et al: Emerging role of quantitative imaging (radiomics) and artificial intelligence in precision oncology. Explor Target Antitumor Ther 2023, 4(4): 569-582.
- Daamen LA, Molenaar IQ, Groot VP: Recent Advances and Future Challenges in Pancreatic Cancer Care: Early Detection, Liquid Biopsies, Precision Medicine and Artificial Intelligence. J Clin Med 2023, 12(23): 7485.
- Jain G, Das P, Ranjan P, Neha, Valderrama F, Cieza-Borrella C: Urinary extracellular vesicles miRNA-A new era of prostate cancer biomarkers. Front Genet 2023, 14: 1065757.
- Owida HA, Hassan MR, Ali AM, Alnaimat F, Al Sharah A, Abuowaida S, Alshdaifat N: The performance of artificial intelligence in prostate magnetic resonance imaging screening. Int J Electr Comput Eng 2024, 14(2): 2234-2241.
- Badenhorst A, John J, Perera M, Adam AG: Prostate cancer screening guidelines: To PSA or not to PSA? Wits J Clin Med 2024, 6(2): 103-108.
- Hugosson J, Månsson M, Wallström J, Axcrona U, Carlsson SV, Egevad L, Geterud K, Khatami A, Kohestani K, Pihl CG et al: Prostate Cancer Screening with PSA and MRI Followed by Targeted Biopsy Only. N Engl J Med 2022, 387(23): 2126-2137.
- Kohestani K, Månsson M, Arnsrud Godtman R, Stranne J, Wallström J, Carlsson S et al: The GÖTEBORG prostate cancer screening 2 trial: a prospective, randomised, population-based prostate cancer screening trial with prostate-specific antigen testing followed by magnetic resonance imaging of the prostate. Scand J Urol 2021, 55(2): 116-124.
- He M, Cao Y, Chi C, Yang X, Ramin R, Wang S, Yang G, Mukhtorov O, Zhang L, Kazantsev A et al: Research progress on deep learning in magnetic resonance imaging-based diagnosis and treatment of prostate cancer: a review on the current status and perspectives. Front Oncol 2023, 13: 1189370.
- Wu X, Wang Y, Yang Q, Thorley N, Punwani S, Kasivisvanathan V, Bonmati E, Hu Y: AI-assisted prostate cancer detection and localisation on biparametric MR by classifying radiologist-positives. In: Medical Imaging 2025: Computer-Aided Diagnosis: 2025, 134073J: 885-890.
- Okobi TJM, Uhomoibhi, T. O. MD., Okobi, O. E. MD., Onyebuchi, E. C. MBBS., Egberuare, E. O. MD., Izundu et al: Screening Asymptomatic Men for Prostate Cancer Using Prostate-Specific Antigen as An Early Detection Tool: A Review of Existing Literature. Int J Sci Adv 2023, 4(1): 8-14.
- Remmers S, Roobol MJ: Personalized strategies in population screening for prostate cancer. Int J Cancer 2020, 147(11): 2977-2987.
- Noriega Landa E, Quaye GE, Su X, Badmos S, Holbrook KL, Polascik TJ, Adams ES, Deivasigamani S, Gao Q, Annabi MH et al: Urinary fatty acid biomarkers for prostate cancer detection. PLoS One 2024, 19(2): e0297615.
- Alijaj N, Pavlovic B, Martel P, Rakauskas A, Cesson V, Saba K, Hermanns T, Oechslin P, Veit M, Provenzano M et al: Identification of Urine Biomarkers to Improve Eligibility for Prostate Biopsy and Detect High-Grade Prostate Cancer. Cancers (Basel) 2022, 14(5): 1135.
- Johnston E, Pye H, Bonet-Carne E, Panagiotaki E, Patel D, Galazi M, Heavey S, Carmona L, Freeman A, Trevisan G et al: INNOVATE: A prospective cohort study combining serum and urinary biomarkers with novel diffusion-weighted magnetic resonance imaging for the prediction and characterization of prostate cancer. BMC Cancer 2016, 16(1): 816.
- Sequeira JP, Salta S, Freitas R, López-López R, Díaz-Lagares Á, Henrique R, Jerónimo C: Biomarkers for Pre-Treatment Risk Stratification of Prostate Cancer Patients: A Systematic Review. Cancers (Basel) 2024, 16(7): 1363.
- Bratt O, Auvinen A, Arnsrud Godtman R, Hellström M, Hugosson J, Lilja H, Wallström J, Roobol MJ: Screening for prostate cancer: evidence, ongoing trials, policies and knowledge gaps. BMJ Oncol 2023, 2(1): e000039.
- Boehm BE, York ME, Petrovics G, Kohaar I, Chesnut GT: Biomarkers of Aggressive Prostate Cancer at Diagnosis. Int J Mol Sci 2023, 24(3): 2185.
- Munteanu VC, Munteanu RA, Gulei D, Schitcu VH, Petrut B, Berindan Neagoe I, Achimas Cadariu P, Coman I: PSA Based Biomarkers, Imagistic Techniques and Combined Tests for a Better Diagnostic of Localized Prostate Cancer. Diagnostics (Basel) 2020, 10(10): 806.
- Eyrich NW, Morgan TM, Tosoian JJ: Biomarkers for detection of clinically significant prostate cancer: contemporary clinical data and future directions. Transl Androl Urol 2021, 10(7): 3091-3103.
- Saltman A, Zegar J, Haj-Hamed M, Verma S, Sidana A: Prostate cancer biomarkers and multiparametric MRI: is there a role for both in prostate cancer management? Ther Adv Urol 2021, 13: 1756287221997186.
- Farha MW, Salami SS: Biomarkers for prostate cancer detection and risk stratification. Ther Adv Urol 2022, 14: 17562872221103988.
- Lophatananon A, Muir KR, Gnanapragasam VJ: The efficacy of different biomarkers and endpoints to refine referrals for suspected prostate cancer: the TARGET study (Tiered integrAted tests for eaRly diaGnosis of clinically significant ProstatE Tumours). BMC Med 2024, 22(1): 440.
- Prensner JR, Rubin MA, Wei JT, Chinnaiyan AM: Beyond PSA: the next generation of prostate cancer biomarkers. Sci Transl Med 2012, 4(127): 127rv123.
- Porpiglia F, De Luca S: Prostate cancer biomarkers: new scenarios in the multi-parametric magnetic resonance imaging era. BJU Int 2017, 120(6): 745-746.
- Chen H, Zhou J, Luo J, Wu Y, Qian Y, Shi Y, Qu F, Shi B, Ding J, Cui X et al: Serum multi-cytokines screening identifies TRAIL and IL-10 as probable new biomarkers for prostate health index diagnostic utility adjustment in grey zone aggressive prostate cancer detection: A single-center data in China. Front Immunol 2022, 13: 901176.
- Polymeri E, Kjölhede H, Enqvist O, Ulén J, Poulsen MH, Simonsen JA, Borrelli P, Trägårdh E, Johnsson Å A, Høilund-Carlsen PF et al: Artificial intelligence-based measurements of PET/CT imaging biomarkers are associated with disease-specific survival of high-risk prostate cancer patients. Scand J Urol 2021, 55(6): 427-433.
- Nelson CR, Ekberg J, Fridell K: Prostate cancer detection in screening using magnetic resonance imaging and artificial intelligence. Open Artif Intell J 2020, 6(1): 1-11.
- Würnschimmel C, Chandrasekar T, Hahn L, Esen T, Shariat SF, Tilki D: MRI as a screening tool for prostate cancer: current evidence and future challenges. World J Urol 2023, 41(4): 921-928.
- Harder FN, Weiss K, Amiel T, Peeters JM, Tauber R, Ziegelmayer S, Burian E, Makowski MR, Sauter AP, Gschwend JE et al: Prospectively Accelerated T2-Weighted Imaging of the Prostate by Combining Compressed SENSE and Deep Learning in Patients with Histologically Proven Prostate Cancer. Cancers (Basel) 2022, 14(23): 5741.
- Davoudi F, Moradi A, Becker TM, Lock JG, Abbey B, Fontanarosa D, Haworth A, Clements J, Ecker RC, Batra J: Genomic and Phenotypic Biomarkers for Precision Medicine Guidance in Advanced Prostate Cancer. Curr Treat Options Oncol 2023, 24(10): 1451-1471.
- Bancroft EK, Raghallaigh HN, Page EC, Eeles RA: Updates in Prostate Cancer Research and Screening in Men at Genetically Higher Risk. Curr Genet Med Rep 2021, 9(4): 47-58.
- Benafif S, Kote-Jarai Z, Eeles RA: A Review of Prostate Cancer Genome-Wide Association Studies (GWAS). Cancer Epidemiol Biomarkers Prev 2018, 27(8): 845-857.
- Ni Raghallaigh H, Eeles R: Genetic predisposition to prostate cancer: an update. Fam Cancer 2022, 21(1): 101-114.
- Davis JW: Advances in Prostate Cancer Diagnosis: Triggers for Prostate Biopsy. In: Prostate Cancer - Leading-edge Diagnostic Procedures and Treatments. https://doi.org/10.5772/64402. Epub ahead of print., edn. Edited by Mohan R. Rijeka: IntechOpen; 2016.
- Sud A, Turnbull C, Houlston R: Will polygenic risk scores for cancer ever be clinically useful? NPJ Precis Oncol 2021, 5(1): 40.
- Radice P, Pharoah PD, Peterlongo P: Personalized testing based on polygenic risk score is promising for more efficient population-based screening programs for common oncological diseases. Ann Oncol 2016, 27(3): 369-370.
- Carlsson S, Assel M, Vickers A: Letter to the editor concerning 'Do prostate cancer risk models improve the predictive accuracy of PSA screening? A meta-analysis'. Ann Oncol 2015, 26(5): 1031.
- Resnick MJ, Guzzo TJ: Patient selection essential in optimizing the benefit of radical prostatectomy for patients with organ-confined prostate cancer. Asian J Androl 2011, 13(6): 789-790.
- Cooperberg MR: Re-examining racial disparities in prostate cancer outcomes. J Clin Oncol 2013, 31(24): 2979-2980.
- Bello NA, Miller EC, Cleary KL, Wapner R: Cases in Precision Medicine: A Personalized Approach to Stroke and Cardiovascular Risk Assessment in Women. Ann Intern Med 2019, 171(11): 837-842.
- Killick E, Morgan R, Launchbury F, Bancroft E, Page E, Castro E, Kote-Jarai Z, Aprikian A, Blanco I, Clowes V et al: Role of Engrailed-2 (EN2) as a prostate cancer detection biomarker in genetically high risk men. Sci Rep 2013, 3: 2059.
- Drăgan A, Drăgan A: Novel Insights in Venous Thromboembolism Risk Assessment Methods in Ambulatory Cancer Patients: From the Guidelines to Clinical Practice. Cancers (Basel) 2024, 16(2): 458.
- Akamatsu S, Terada N, Takata R, Kinoshita H, Shimatani K, Momozawa Y, Yamamoto M, Tada H, Kawamorita N, Narita S et al: Clinical Utility of Germline Genetic Testing in Japanese Men Undergoing Prostate Biopsy. JNCI Cancer Spectr 2022, 6(1): pkac001.
- Alqahtani S: Systematic Review of AI-Assisted MRI in Prostate Cancer Diagnosis: Enhancing Accuracy Through Second Opinion Tools. Diagnostics (Basel) 2024, 14(22): 2576.
- Twilt JJ, van Leeuwen KG, Huisman HJ, Fütterer JJ, de Rooij M: Artificial Intelligence Based Algorithms for Prostate Cancer Classification and Detection on Magnetic Resonance Imaging: A Narrative Review. Diagnostics (Basel) 2021, 11(6): 959.
- Patel HD, Remmers S, Ellis JL, Li EV, Roobol MJ, Fang AM, Davik P, Rais-Bahrami S, Murphy AB, Ross AE et al: Comparison of Magnetic Resonance Imaging-Based Risk Calculators to Predict Prostate Cancer Risk. JAMA Netw Open 2024, 7(3): e241516.
- Guerra A, Alves FC, Maes K, Joniau S, Cassis J, Maio R, Cravo M, Mouriño H: Early biomarkers of extracapsular extension of prostate cancer using MRI-derived semantic features. Cancer Imaging 2022, 22(1): 74.
- Guerra A, Alves FC, Maes K, Maio R, Villeirs G, Mouriño H: Risk Biomarkers for Biochemical Recurrence after Radical Prostatectomy for Prostate Cancer Using Clinical and MRI-Derived Semantic Features. Cancers (Basel) 2023, 15(21): 5296.
- Nam R, Patel C, Milot L, Hird A, Wallis C, Macinnis P, Singh M, Emmenegger U, Sherman C, Haider MA: Prostate MRI versus PSA screening for prostate cancer detection (the MVP Study): a randomised clinical trial. BMJ Open 2022, 12(11): e059482.
- Sun Z, Wang K, Kong Z, Xing Z, Chen Y, Luo N, Yu Y, Song B, Wu P, Wang X et al: A multicenter study of artificial intelligence-aided software for detecting visible clinically significant prostate cancer on mpMRI. Insights Imaging 2023, 14(1): 72.
- Bao J, Qiao X, Song Y, Su Y, Ji L, Shen J, Yang G, Shen H, Wang X, Hu C: Prediction of clinically significant prostate cancer using radiomics models in real-world clinical practice: a retrospective multicenter study. Insights Imaging 2024, 15(1): 68.
- Merriel SWD, Buttle P, Price SJ, Burns-Cox N, Walter FM, Hamilton W, Spencer AE: Early economic evaluation of magnetic resonance imaging for prostate cancer detection in primary care. BJUI Compass 2024, 5(9): 855-864.
- Hao S, Discacciati A, Eklund M, Heintz E, Östensson E, Elfström KM, Clements MS, Nordström T: Cost-effectiveness of Prostate Cancer Screening Using Magnetic Resonance Imaging or Standard Biopsy Based on the STHLM3-MRI Study. JAMA Oncol 2022, 9(1): 88-94.
- Huh JS, Park KK: A cost-benefit comparison of biparametric magnetic resonance imaging versus conventional prostate cancer screening. J Urol Oncol 2023, 21(2): 121-127.
- Sandeman K, Blom S, Koponen V, Manninen A, Juhila J, Rannikko A, Ropponen T, Mirtti T: AI Model for Prostate Biopsies Predicts Cancer Survival. Diagnostics (Basel) 2022, 12(5): 1031.
- Pizurica M, Larmuseau M, Van der Eecken K, de Schaetzen van Brienen L, Carrillo-Perez F, Isphording S, Lumen N, Van Dorpe J, Ost P, Verbeke S et al: Whole Slide Imaging-Based Prediction of TP53 Mutations Identifies an Aggressive Disease Phenotype in Prostate Cancer. Cancer Res 2023, 83(17): 2970-2984.
- Dominguez I, Rios-Ibacache O, Caprile P, Gonzalez J, San Francisco IF, Besa C: MRI-Based Surrogate Imaging Markers of Aggressiveness in Prostate Cancer: Development of a Machine Learning Model Based on Radiomic Features. Diagnostics (Basel) 2023, 13(17): 2779.
- Ferro M, de Cobelli O, Vartolomei MD, Lucarelli G, Crocetto F, Barone B, Sciarra A, Del Giudice F, Muto M, Maggi M et al: Prostate Cancer Radiogenomics-From Imaging to Molecular Characterization. Int J Mol Sci 2021, 22(18): 9971.
- Li G, Tholance Y, Mallouk N, Waeckel L, Flandrin P, Bali B, Badet L, Cornillon P: Quantification of Urinary Exosomal Prostate-Specific Antigen for the Diagnosis of Prostate Cancer Using Clinical Laboratory-Based Techniques: Protocol for a Case-Control Study. JMIR Res Protoc 2024, 13: e63551.
- de la Calle C, Fasulo V, Maggi M, Buffi N, Cooperberg M, Carroll P, Shinohara K, Nguyen H: MP30-11 clinical utility of exosomedx when combined with psa, trus and mpmri for the detection of high-grade prostate cancer. J Urol 2021, 206(Supplement 3): e503-e504.
- Lazzeri M, Fasulo V, Tinterri C: Eve's and Adam's rib for prostate cancer screening. BJU Int 2023, 131(6): 637-638.
- Grollemund V, Pradat PF, Querin G, Delbot F, Le Chat G, Pradat-Peyre JF, Bede P: Machine Learning in Amyotrophic Lateral Sclerosis: Achievements, Pitfalls, and Future Directions. Front Neurosci 2019, 13: 135.
- Thompson LI, Strenger JR, Emrani S, De Vito A, Harrington K, Roque N, Salloway S, Sliwinski M, Correia S: 91 Remote Smartphone-Based Assessment Predicts Standard Neuropsychological Test Performance and Cerebral Amyloid Status in Cognitively Normal Older Adults. J Int Neuropsychol Soc 2023, 29(s1): 493-494.
- Mizuno K, Beltran H: Future directions for precision oncology in prostate cancer. Prostate 2022, 82 Suppl 1(Suppl 1): S86-S96.
- Cho HR, Jeon H, Park CK, Park SH, Kang KM, Choi SH: BCAT1 is a New MR Imaging-related Biomarker for Prognosis Prediction in IDH1-wildtype Glioblastoma Patients. Sci Rep 2017, 7(1): 17740.
- Arthur A, Johnston EW, Winfield JM, Blackledge MD, Jones RL, Huang PH, Messiou C: Virtual Biopsy in Soft Tissue Sarcoma. How Close Are We? Front Oncol 2022, 12: 892620.
- Zetterberg H, Burnham SC: Blood-based molecular biomarkers for Alzheimer's disease. Mol Brain 2019, 12(1): 26.
- Matulewicz RS, Fankhauser CD, Sheinfeld J, Bagrodia A: Novel approaches to redesign surveillance strategies following orchiectomy for localized testicular cancer: a narrative review. Transl Androl Urol 2023, 12(6): 1016-1022.
- Esen T, Turkbey B, Patel A, Futterer J: Multiparametric MRI in prostate cancer. Biomed Res Int 2014, 2014: 296810.
- Tawadros T, Valerio M: Addressing overtreatment following the diagnosis of localized prostate cancer. Expert Rev Anticancer Ther 2016, 16(4): 373-374.
- Joniau S: Introduction. Dilemmas in the field of prostate cancer detection and treatment. Acta Oncol 2011, 50 Suppl 1: 24.
- Lüdemann C, Reinersmann J-L, Klinger C, Degener S, Dreger NM, Roth S, Kaufmann M, Savelsbergh A: Prostate Cancer Specific Exosomal miRNAs in Saliva: a Way to a New and Dependable Screening Method. 2020 Epub ahead of print.
- Farahani H, Alaee M, Amri J, Baghinia MR, Rafiee M: Serum and Saliva Concentrations of Biochemical Parameters in Men with Prostate Cancer and Benign Prostate Hyperplasia. Lab Med 2020, 51(3): 243-251.
- Santaolalla A, Hulsen T, Davis J, Ahmed HU, Moore CM, Punwani S, Attard G, McCartan N, Emberton M, Coolen A et al: The ReIMAGINE Multimodal Warehouse: Using Artificial Intelligence for Accurate Risk Stratification of Prostate Cancer. Front Artif Intell 2021, 4: 769582.
- Schulman AA, Polascik TJ: Most of patients with localized prostate cancer will be treated in the future? Opinion: No. Int Braz J Urol 2017, 43(4): 584-587.
- Nikles S, Pezelj I, Tomić M, Knežević M, Vrhovec B, Dumbović L, Pirša M, Kavelj I, Tomašković I: Current role of magnetic resonance imaging in the screening, diagnosis, and treatment of prostate cancer. Acta clinica Croatica 2022, 61(Supplement 3): 92-94.
- Sanghera S, Coast J, Martin RM, Donovan JL, Mohiuddin S: Cost-effectiveness of prostate cancer screening: a systematic review of decision-analytical models. BMC Cancer 2018, 18(1): 84.
- Lazareva O, Riediger A, Stegle O, Sueltmann H, Hohenfellner M, Goertz M: Integration of serum androgens and Sex Hormone-Binding Globulin for optimized early detection of aggressive prostate cancer. medRxiv 2024, 12.05.24318544.
Annals of urologic oncology
p-ISSN: 2617-7765, e-ISSN: 2617-7773
 Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript