Review Article | Open Access
The Recent Research Progress of CircRNA in Bladder Cancer
Shuo Liu1, Xin-liang Xu21Jining center for disease control and prevention, Jining, 272011, China.
2Department of Pain, Jining No.1 Peoples Hospital, Jining, 272011, China.
Correspondence: Xin-liang Xu (Department of Pain, Jining No.1 Peoples Hospital, Jining, 272011, China; Email:2466082197@qq.com ).
Annals of Urologic Oncology 2023, 6(1): 10-17. https://doi.org/10.32948/auo.2023.02.09
Received: 04 Feb 2023 | Accepted: 09 Feb 2023 | Published online: 10 Feb 2023
Key words circRNA, miRNA, ceRNA, bladder cancer
Bladder cancer (BC) refers to malignant tumor(s) that occurs on the bladder mucosa, and is mainly divided into three types: urinary epithelial cancer, squamous carcinoma, and adenocarcinoma. The highest percentage is urinary epithelial cancer, accounts for about 90% of total bladder cancer [1, 6]. According to statistics, bladder cancer is one of the most common malignant tumors in the urogenital system. It is ninth in position among the common cancer incidence in the world. It is the thirteenth for the cause of cancer-related death [7]. In China, the incidence and mortality of bladder cancer is first among the urinary system tumors, and with the advancement and changes in the lifestyle habits, the incidence and detection rate of bladder cancer has been increasing year by year [7, 8].
Diagnosis and treatment methods for bladder cancer in clinic
At present, there are numerous screening methods for bladder cancer clinically, such as urine cytological examinations, optical imaging, tumor marker testing, imaging examination, cystoscopy, biopsy, diagnostic transurethral resection of bladder tumor (TURBT) with surgery pathological examinations [9]. Among them, cystoscopy is a gold standard for the diagnosis of bladder cancer, and is very important especially before the resection of bladder tumor, which is essential for the complete resection of the bladder tumor during surgery. However, standard white light cystoscopy might miss up to 20% of bladder tumors [10]. Most common treatment methods for bladder cancer are surgical resection, which mainly include TURBT, radical cystectomy, bladder perfusion therapy, bladder irrigation immunotherapy etc. For poorly differentiated and partial well-differentiated bladder urinary epithelial cancer, TURBT remains the first choice. However, the recurrence rate within one year was 50% for high-risk bladder cancer after surgery, and the recurrence rate within 5 years after surgery remains high up to 90% [11, 12]. Therefore, a problem exists due to the long treatment duration and high postoperative recurrence rate. High interest in recent years and focus remains on fast, and accurate bladder cancer screening and diagnosis methods. Among them, the abnormal expression of circRNA may play an important role in the development of bladder cancer, and has certain application prospects in the diagnosis and treatment of bladder cancer.
The Sanger team discovered a new RNA molecule in the virus in 1976, which was named CircRNA [13]. CircRNA is a category of closed single-chain with covalent and without 5’-hats and 3’-polya tails. It is composed of exon and/or intron, which is not degraded by RNA exonuclease. It is one of the non-coded RNAs, and existed in the cells, serum exogenous bodies, and saliva having a huge potential for regulatory control [14, 15]. Researches found that CircRNA participated in the occurrence and development of cancer, high stability, and evolution conservative characteristics [16].
CircRNA biological occurrence and mechanism
CircRNA organisms mainly include four models: Splice looping of the exon lariat, Looping of intron pairs, Splice looping of the inner ring daughter lariat, Splice looping of internal ring rope, cyclization driven by RNA-binding proteins or trans factors [17]. Most circRNAs are encoded by known protein-coding genes and fall into three categories based on their structural composition: exon circRNA(EcircRNA), annular intron RNA(ciRNA), and exon-intron circRNA(EIciRNA) [18].
CircRNA plays different biological functions in different biological processes, mainly including five aspects [19]: (1) circRNA can be used as "molecular sponge", which is competitively combined with some miRNA (microRNA). To a certain extent, inhibitory effect of miRNA on mRNA indirectly promotes the expression of the target mRNA; (2) The complexes of circRNA combining with small ribonucleoprotein particles (snRNP) can further interact with RNA polymerase, to regulate the gene transcription; (3) CircRNA can interact with certain proteins to affect its function; (4) Some CircRNAs can translate into protein and performed some or other functions of the host gene; (5) CircRNA is a single-chain ring structure, which can form a dual chain structure with some mRNAs, thereby adjusting the expression of mRNA. Among them, circRNA as a "molecular sponge" combines some miRNA to promote the expression of target mRNA [20]. This is regarded as competing endogenous RNAS (ceRNA) early, that is, the ceRNA mechanism. In the ceRNA mechanism, circRNA and mRNA, which have a binding site with the same miRNA, have a positive adjustment relationship, which can indirectly predict the expression trend of target mRNA through the expression trend of circRNA, and vice versa [21-23].
Figure 1 shows the circRNA classification and mechanism of circRNA acting as a miRNA molecular sponge.
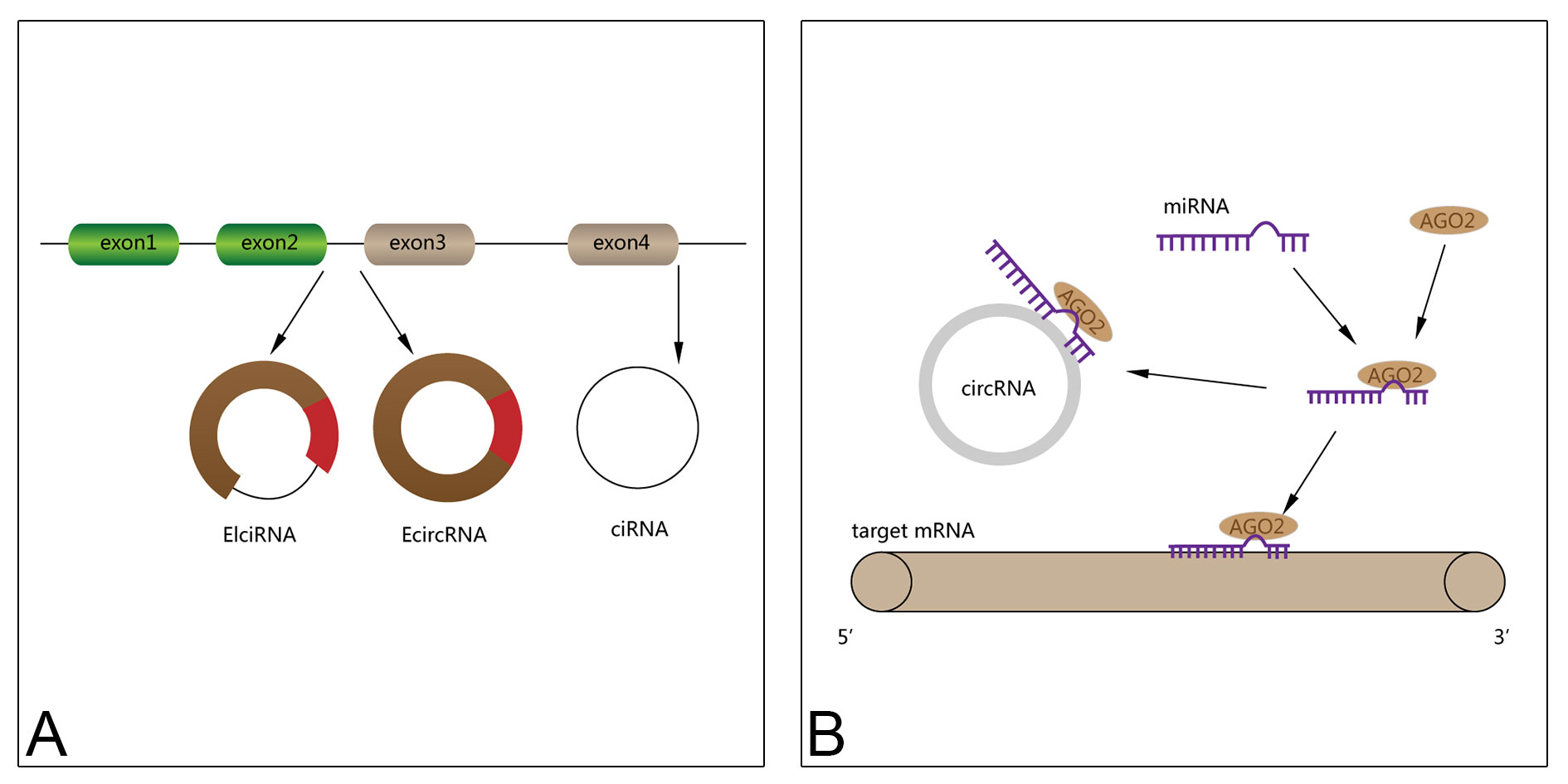 Figure 1. Schematic diagram for circRNA classification (A) and mechanism of circRNA acting as a miRNA molecular sponge (B).
Figure 1. Schematic diagram for circRNA classification (A) and mechanism of circRNA acting as a miRNA molecular sponge (B).
Figure 2 shows that circRNAs regulate cell proliferation, apoptosis, invasion, migration and metastasis, angiogenesis, and cisplatin chemoresistance in BCa cells.
Study of partial tumor suppressor circRNA in bladder cancer
circHIPK3. circBCRC-2 (bladder cancer-related circular RNA-2, circBCRC-2), also known as circHIPK3, is a ring-shaped RNA that is mainly existed in the cytoplasm, which is formed by splicing of exons of the HIPK3 gene. In vitro, the excessive expression can effectively inhibit the migration and invasion of the bladder cancer cells. Its low expression level positively correlate with highly grade tumors and lymphatic metastasis in bladder cancer. circHIPK3 mainly utilizes the expression of heparanase (HPSE), which is used as a "molecular sponge", and plays the role of cancer suppressor gene in bladder cancer [24, 25].
circBCRC-3. circBCRC-3 (bladder cancer-related circular RNA-3, circBCRC-3) is a ring-shaped RNA derived from the PSMD1 gene. Over-expression can inhibit the proliferation of bladder cancer cells. circBCRC-3 mainly promotes the expression of the P27 through the "molecular sponge" as the "molecular sponge" of the MIR -182-5P and plays the role of cancer suppressor gene [24-27].
circUBXN7. circUBXN7 (hsa_circ_001380) is a circular RNA that expresses significantly down in the bladder cancer tissue and cell lines, and its lower level is positively related with bladder cancer staging, grading, prognosis. Excessive expression of circUBXN7 in vitro can significantly inhibit the cell proliferation, migration and invasion. Excessive expression of cancer cells in the body can inhibit tumor growth. circUBXN7 is mainly used as the expression of recombinant human beta-1, 4 -galactosyltransferase3 (B4GALT3) to play a role of cancer suppressor gene [28].
circSLC8A1. circSLC8A1 is a ring-shaped RNA that is significantly downregulated in bladder cancer tissue, which can inhibit proliferation, migration and invasion of bladder cancer cells. Its low expression is positively associated with bladder cancer, which are mainly used as ‘molecular sponge’ of miR-494 to adjust the expression of PTEN and plays the role of cancer [29].
circITCH. circITCH is the famous circular RNA that is formed by splicing of ITCH (itchy E3 ubikuitin protein ligase) gene exon. The expression of circITCH is significantly lowered in bladder cancer tissue and cell lines. Its excessive expression inhibits proliferation, migration, and invasion in bladder cancer cells. High expression of circITCH also results in tumor growth and metastasis, and its expression level is proportional to the healing survival time in the bladder cancer. circITCH adjusts the suppressor genes role of p21 and PTEN (P21 and phosphatase tensin homologue) in bladder cancer, which mainly through the "molecular sponge" of MIR -17 and MIR -24 [30].
circACVR2A. circACVR2A (HSA_CIRC_ 001073) is a ring-shaped RNA that is mainly located in the cytoplasm. It is expressed at lower level in the bladder cancer tissue and cell lines. Overexpression of circACVR2A significantly inhibits proliferation, migration and invasion of bladder cancer cells. In vivo, overexpression can inhibit tumor growth and lymphatic metastasis, and its low expression has a certain correlation with poor healing. circACVR2A is mainly targeted by the "molecular sponge" as the mir-626 (eyes absent 4, EYA4), thereby playing the role of cancer suppressor gene [31].
circPICALM. circPICALM is mainly from the cell transcripts of PICALM gene exon 9 to 12. Its expression is downregulated in bladder cancer tissue and cancer cell lines. Overexpression of circPICALM results in significant inhibition in the migration and invasion ability of bladder cancer cells. Its low expression positively correlate with the pathological staging, classification, and lymph node metastasis of bladder tumors. It also has a certain correlation with overall survival of patients with bladder cancer. It is projected as a putative prognostic biomarker before bladder cancer resection. circPICALM mainly regulates the expression of STEAP4 (six transmembrane epithelial antigen of the prostate 4, STEAP4) by ‘molecular sponge’ of MIR-1265, and play a role in the role of cancer suppressor gene in bladder cancer [32].
circZKSCAN1. circZKSCAN1 is a ring-shaped RNA, which is mainly located in the cytoplasm. It expression is lowered in bladder cancer tissue and cell lines. Forced expression can inhibit proliferation, migration and invasion of bladder cancer cells, however its lower expression is positively associated with poor tumor survival rate, high recurrence, and positive tumor metastasis. circZKSCAN1 mainly regulates the expression of p21 through the ‘molecular sponge’ as miR -1178-3P, and play a role in the cancer suppressor gene in bladder cancer [33].
circMTO1. circMTO1 is an important circRNA frequently downregulated in bladder cancer tissue. Lower circMTO1 expression positively correlate with bladder cancer metastasis and poor survival. It was shown that circMTO1 was able to sponge miR-221 and overexpression of circMTO1 negatively regulate the E-cadherin/N-cadherin pathway to inhibit bladder cancer cells EMT by competing for miR-221 [34].
NR3C1. It was demonstrated that circNR3C1 possessed four targeting sites of miR-27a-3p and could effectively sponge miR-27a-3p to suppress the expression of cyclin D1, which can inhibit cell cycle progression and proliferation of bladder cancer cells [35].
CRIM1. circCRIM1could effectively sponge miR-182 to suppress the expression of Foxo3a, which can inhibit invasion and migration of cancer cells to some extent [36].
Study of cancer-promoting circRNAs in bladder cancer
circVANGL1. CIRCVANGL1 (HSA_CIRC_002623) is a ring-shaped RNA and its expression is high in the bladder cancer tissue and cell lines. Loss of expression of VANGL1 in vitro can inhibit the proliferation, migration, and invasion of bladder cancer cells. Mechanistically, circVANGL1 promotes IGFBP2 (insulin like growth factor binding protein 2, IGFBP2) to promote the role of cancer genes through the miR-114 ‘molecular sponge’ [37].
circUVRAG. circUVRAG is a ring-shaped RNA mostly found in cytoplasm with high expression in bladder cancer cell lines. Knockdown of circUVRAG in bladder cancer cells resulted in the inhibition of tumor growth, metastasis in mouse xenograft model. circUVRAG is essentially used as the ‘molecular sponge’ of miR-223 to adjust FGFR2 (Fibroblast growth factor receptor 2, FGFR2) [38].
circTFRC. circTFRC is a ring-shaped RNA expressed in the bladder cancer tissue and cell lines. High expression of circTFRC promote proliferation, invasion and induction of epithelial-mesenchymal transition (EMT) of bladder cancer cells; whereas higher levels positively correlate with the s tumor grading, T staging and lymphatic metastasis in bladder cancer patients. circTFRC regulates the role of TFRC (transferin receptor, TFRC) through the ‘molecular sponge’ of MIR-107, and play a role in oncogenes in bladder cancer [39].
circBPTF. circBPTF (hsa_circ_0000799) is a ring-shaped RNA from the reverse editing of the precursor gene BPTF (Bromodomain PHD finger transcription factor, BPTF). It is located in the cytoplasm of tissues and cell lines. Over-expression of circBPTF significantly promotes migration of bladder cancer cells and high circBPTF expression is associated with poor survival, higher tumor grade and tumor recurrence rate of patients with bladder cancer. circBPTF regulates the role of Rab27 (Ras -Related protein Rab-27A, RAB27A) through the ‘molecular sponge’ of miR-31-5p [40].
circPRMT5. circPRMT5 is a ring-shaped RNA increased in cancer tissue, serum, and urinary exosomes of the patients with urothelial carcinoma. circPRMT5 positively correlates with poor survival such as advanced clinical stage of patients and the decrease in/or decreased survival rate, which can predict the lymphatic metastasis. circPRMT5 indirectly adjusts SNAIL1 (recombinant snail homolog 1) by ‘molecular sponge’ of MIR-30C. A study found that the expression of circPRMT5 is not significantly increased in other human cancers. The result indicates that circPRMT5 expression in urothelial carcinoma may be tumor-specific. Therefore, circPRMT5 may become a specific biomarker that predicts lymphatic metastasis in urothelial carcinoma [41].
circRNA-MYLK. circRNA-MYLK and VEGFA have been shown to significantly upregulate and co-expressed in bladder cancer. circRNA-MYLK levels were related to the progression stage and grade of bladder cancer. circRNA-MYLK could directly bind to miR-29a and release the suppression for target VEGFA, which activates VEGFA/VEGFR2 signaling pathway. Increased expressing circRNA-MYLK accelerated cell proliferation, migration, tube formation of HUVEC and rearranged cytoskeleton. In addition, up-regulating circRNA-MYLK promoted epithelial-mesenchymal transition (EMT) [42].
Circular RNA CEP128. circCEP128 acts as a ceRNA for miR-145-5p, which could upregulate SOX11 and further promotes cell proliferation and inhibits apoptosis of bladder cancer cells [43, 44].
RIP2. In vitro and in vivo studies suggest that circRIP2 enables to promote bladder cancer progression via inducing EMT. It was found that circRIP2 enables to sponge miR-1305 to elevate Tgf-β2 in bladder cancer, and inducing EMT via TGF-β2/smad3 pathway [45].
INTS4. circINTS4 was found to control multiple pathological processes, including cell proliferation and migration, cell cycle and apoptosis promoting bladder tumorigenesis. circINTS4 directly binds to miR-146b to inhibit its activity by targeting 3'-UTR of CARMA3 mRNA. In addition, circINTS4 could activate the NF-kB signaling pathway and suppress the p38-MAPK signaling pathway in a CARMA3-mediated manner in bladder cancer cells [46].
ZNF139. Bioinformatics research suggests that ZNF139/circZNF139 facilitates its effects on the proliferation, clonal expansion, migration, and invasive potential of bladder cancer cells [47].
KDM4C. circKDM4C directly interacts with miR-200b-3p and miR-200c-3p as a miRNA sponge, enhancing the expression of ZEB1 and promotes mesenchymal phenotype [48].
EHBP1. Mechanistically, circEHBP1 overexpression promote TGFbR1 expression by attenuating miR-130a-3p to activate the TGF-b/SMAD3 signaling pathway, further elevating VEGF-D secretion and ultimately facilitating lymphangiogenesis and lymph node metastasis in bladder cancer [49].
ZNF609. In bladder cancer patients, increased expression of circZNF609 correlate with poor survival. In vitro and in vivo, forced expression of circZNF609 enhance bladder cancer cell proliferation, migration, and cisplatin chemo-resistance. Mechanistically, circZNF609 alleviated the inhibition effect on target CDC25B expression by sponging miR-1200 [50].
SPECC1. Increased expression of circSPECC1 contribute to poor prognosis of bladder cancer. Knockdown of circSPECC1 impairs proliferation and migration of bladder cancer cells. Mechanically, circSPECC1 sponge miR-136-5p to promote the mRNA and protein expression of GNAS [51].
NT5E. Decreased miR-502-5p reversed the circNT5E silencing-mediated inhibition of bladder cancer cell growth and migration. The study suggested that circNT5E may act as a pro-oncogene in the development and progression of bladder cancer and it may become a useful tumor biomarker and promising therapeutic target for treatment [52].
|
Table 1. Summary on circRNA associated with cancer. |
|||||
|
Classification |
circRNA |
ceRNA |
Target gene |
Function |
References |
|
Tumor supressor circRNAs |
HIPK3 |
miR-588 |
HPSE |
To inhibit cancer cell migration, invasion and angiogenesis |
[24, 25] |
|
BCRC3 |
miR-182-5p |
p27 |
To inhibit the growth and proliferation of cancer cells and induced G0/ G1 phase arrest |
[24-27] |
|
|
UBXN7 |
miR-1247-3P |
B4GALT3 |
To inbibit cancer cell proliferation, migration, invasion and tumor growth |
[28] |
|
|
SLC8A1 |
miR-130b,miR-494 |
PTEN |
To inhibit cancer cell proliferation, migration, invasion and tumor growth |
[29] |
|
|
ITCH |
miR-17/miR-224 |
P21/PTEN |
To inhibit cancer cell proliferation, migration, invasion, tumor growth and metastasis |
[30] |
|
|
ACVR2A |
miR-626 |
EYA4 |
To inhibit cancer cell proliferation, migration, invasion, tumor growth, lymphatic metastasis |
[31] |
|
|
PICALM |
miR-1265 |
STEAP4 |
To inhibit cancer cell migration, invasion and lymphatic metastasis |
[32] |
|
|
ZKSCAN1 |
miR-1178-3P |
p21 |
To inhibit the proliferation, migration and invasion of cancer cells and induced G1/ S phase arrest |
[33] |
|
|
MTO1 |
miR-221 |
E-cadherin/N-cadherin pathway |
To inhibit bladder cancer cells' EMT by competing for miR-221 |
[34] |
|
|
NR3C1 |
miR-27a-3p |
cyclin D1 |
To inhibit cell cycle progression and proliferation of bladder cancer cells in vitro, as well as suppress tumor growth in vivo |
[35] |
|
|
Onco-circRNAs |
VANGL1 |
miR-1184 |
IGFBP2 |
To promote the proliferation, migration and invasion of cancer cells |
[36] |
|
UVRAG |
miR-223 |
FGFR2 |
To promote cancer cell proliferation, metastasis, tumor growth |
[37] |
|
|
TFRC |
miR-107 |
TFRC |
To promote cancer cell proliferation, invasion, lymphatic metastasis, and induces EMT of bladder cancer cells |
[38] |
|
|
BPTF |
miR-31-5p |
RAB27A |
To promote cancer cell migration and invasion |
[39] |
|
|
PRMT5 |
miR-30c |
SNAIL1 |
To promote cancer cell migration, invasion, lymphatic metastasis, and induces EMT of bladder urothelial cancer cells |
[40] |
|
|
MYLK |
miR-29a |
VEGFA |
To accelerate cell proliferation, migration, tube formation of HUVEC and rearranged cytoskeleton |
[41] |
|
|
CEP128 |
miR-145-5p |
SOX11 |
To further promote cell proliferation and inhibits cell apoptosis of bladder cancer |
[42, 43] |
|
|
RIP2 |
miR-1305 |
TGF-β2 |
Effective circRIP2 activity accelerates bladder cancer progression via inducing EMT |
[44] |
|
|
INTS4 |
miR-146b |
CARMA3 |
To control multiple pathological processes, including cell proliferation and migration, the cell cycle and apoptosis to promotes tumorigenesis in bladder cancer |
[45] |
|
|
ZNF139 |
- |
PI3K/AKT pathway |
To promote cell proliferation, migration and invasion via activation of PI3K/AKT pathway in bladder cancer |
[46] |
|
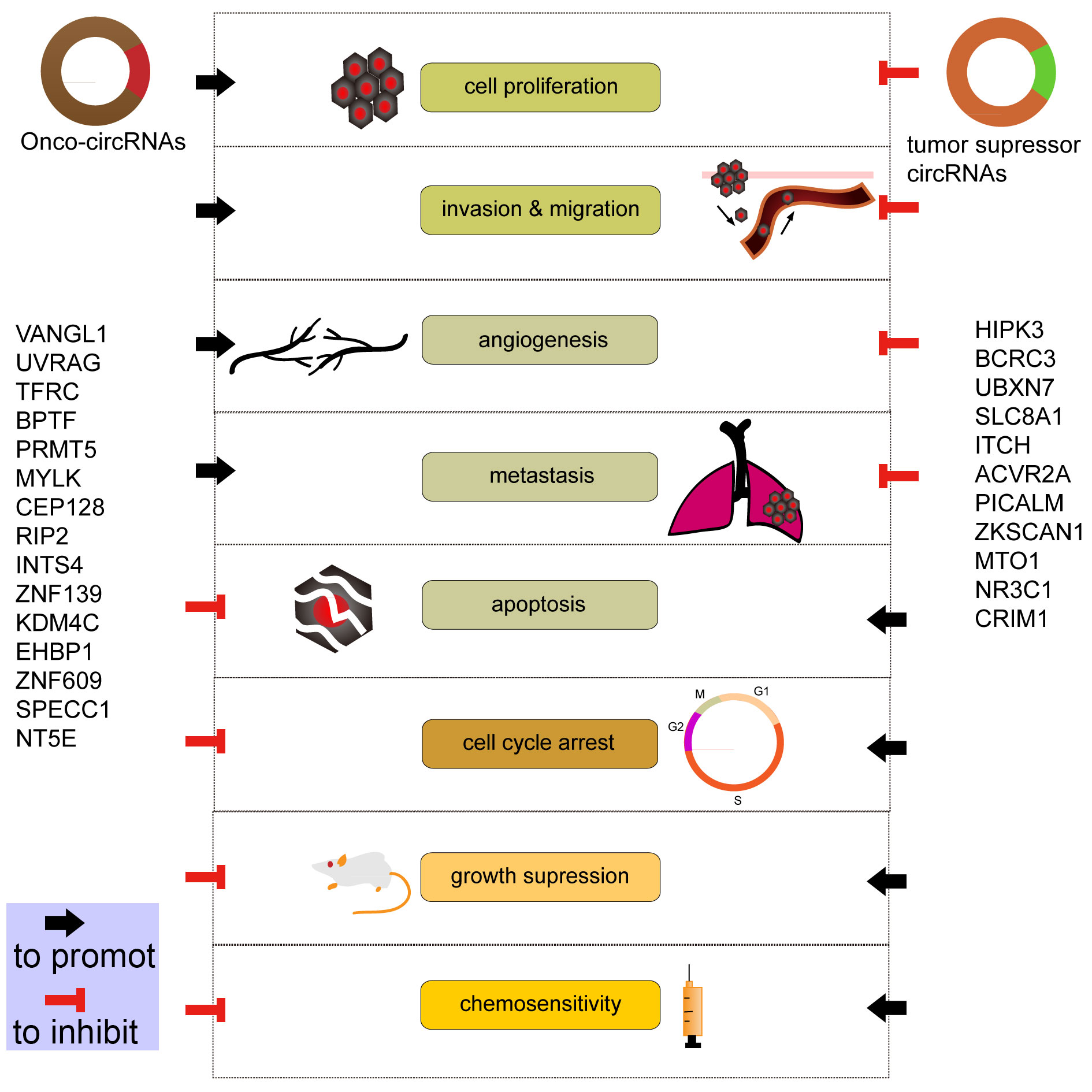 Figure 2. circRNAs regulate cell proliferation, apoptosis, invasion, migration and metastasis, angiogenesis, and cisplatin chemoresistance in BCa cells.
Figure 2. circRNAs regulate cell proliferation, apoptosis, invasion, migration and metastasis, angiogenesis, and cisplatin chemoresistance in BCa cells.
We thank Dr. Sanjay Gupta for their proofreading on the manuscript.
Ethical policy
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study. Approval from institutional ethical committee was taken.
Availability of data and materials
All data generated or analysed during this study are included in this publication.
Author contributions
SL designed the study and was responsible for the writing of the original draft. XLX and SL edited and approved the final manuscript.
Competing interests
All authors declare no competing interests.
Funding
This study was supported by Traditional Chinese Medicine Science and Technology Project of Shandong Province (2021M079).
- Lobo N, Afferi L, Moschini M, Mostafid H, Porten S, Psutka SP, Gupta S, Smith AB, Williams SB, Lotan Y: Epidemiology, Screening, and Prevention of Bladder Cancer. Eur Urol Oncol 2022, 5(6): 628-639.
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F et al: Landscape of transcription in human cells. Nature 2012, 489(7414):101-108.
- Li Y, Li G, Guo X, Yao H, Wang G, Li C: Non-coding RNA in bladder cancer. Cancer Lett 2020, 485: 38-44.
- Wang P, Wang Z, Zhu L, Sun Y, Castellano L, Stebbing J, Yu Z, Peng L: A pyroptosis-related lncRNA signature in bladder cancer. Cancer Med 2022, https://doi.org/10.1002/cam4.5344. Epub ahead of print.
- Li K, Yao T, Wang Z: lncRNA-mediated ceRNA network in bladder cancer. Noncoding RNA Res 2023, 8(2): 135-145.
- Schafer EJ, Jemal A, Wiese D, Sung H, Kratzer TB, Islami F, Dahut WL, Knudsen KE: Disparities and Trends in Genitourinary Cancer Incidence and Mortality in the USA. Eur Urol 2022, https://doi.org/10.1016/j.eururo.2022.11.023. Epub ahead of print.
- DePalo DK, Elleson KM, Carr MJ, Spiess PE, Zager JS: Genitourinary melanoma: An overview for the clinician. Asian J Urol 2022, 9(4): 407-422.
- Xiang Z, Ye Z, Ma J, Lin Y, Zhou Y: Temporal Trends and Projections of Bladder Cancer Burden in China from 1990 to 2030: Findings from the Global Burden of Disease Study. Clin Epidemiol 2022, 14: 1305-1315.
- Clark KR: Bladder Cancer: Types, Risk Factors, Diagnosis, and Treatment. Radiol Technol 2022, 94(1): 46-50.
- Herr HW: Randomized trial of narrow-band versus white-light cystoscopy for restaging (second-look) transurethral resection of bladder tumors. Eur Urol 2015, 67(4): 605-608.
- Jobczyk M, Stawiski K, Fendler W, Rozanski W: Validation of EORTC, CUETO, and EAU risk stratification in prediction of recurrence, progression, and death of patients with initially non-muscle-invasive bladder cancer (NMIBC): A cohort analysis. Cancer Med 2020, 9(11): 4014-4025.
- Lin X, Deng T, Wu S, Lin SX, Wang D, Wu CL: The clinicopathological characteristics and prognostic value of squamous differentiation in patients with bladder urothelial carcinoma: a meta-analysis. World J Urol 2020, 38(2): 323-333.
- Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK: Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976, 73(11): 3852-3856.
- Wilusz JE, Sharp PA: Molecular biology. A circuitous route to noncoding RNA. Science 2013, 340(6131): 440-441.
- Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S: Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015, 25(8): 981-984.
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M et al: Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495(7441): 333-338.
- Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H: Circular RNA: A new star of noncoding RNAs. Cancer Lett 2015, 365(2): 141-148.
- Ebbesen KK, Kjems J, Hansen TB: Circular RNAs: Identification, biogenesis and function. Biochim Biophys Acta 2016, 1859(1): 163-168.
- Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M: CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 2017, 16(1): 94.
- Zhou J, Qiu C, Fan Z, Liu T, Liu T: Circular RNAs in stem cell differentiation: a sponge-like role for miRNAs. Int J Med Sci 2021, 18(11): 2438-2448.
- An Y, Furber KL, Ji S: Pseudogenes regulate parental gene expression via ceRNA network. J Cell Mol Med 2017, 21(1):185-192.
- Sen R, Ghosal S, Das S, Balti S, Chakrabarti J: Competing endogenous RNA: the key to posttranscriptional regulation. ScientificWorldJournal 2014, 2014: 896206.
- Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W: ceRNA in cancer: possible functions and clinical implications. J Med Genet 2015, 52(10): 710-718.
- Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, Liu D, Wang M, Wang L, Zeng F et al: CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep 2017, 18(9): 1646-1659.
- Xie F, Li Y, Wang M, Huang C, Tao D, Zheng F, Zhang H, Zeng F, Xiao X, Jiang G: Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol Cancer 2018, 17(1): 144.
- Li B, Xie F, Zheng FX, Jiang GS, Zeng FQ, Xiao XY: Overexpression of CircRNA BCRC4 regulates cell apoptosis and MicroRNA-101/EZH2 signaling in bladder cancer. J Huazhong Univ Sci Technolog Med Sci 2017, 37(6): 886-890.
- Gu C, Zhao K, Zhou N, Liu F, Xie F, Yu S, Feng Y, Chen L, Yang J, Tian F et al: UBAC2 promotes bladder cancer proliferation through BCRC-3/miRNA-182-5p/p27 axis. Cell Death Dis 2020, 11(9): 733.
- Liu H, Chen D, Bi J, Han J, Yang M, Dong W, Lin T, Huang J: Circular RNA circUBXN7 represses cell growth and invasion by sponging miR-1247-3p to enhance B4GALT3 expression in bladder cancer. Aging (Albany NY) 2018, 10(10): 2606-2623.
- Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen W, Jiang B, Qin H, Guo X, Liu M et al: Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer 2019, 18(1): 111.
- Yang C, Yuan W, Yang X, Li P, Wang J, Han J, Tao J, Li P, Yang H, Lv Q et al: Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer 2018, 17(1): 19.
- Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q, Wang Q, Xie R, Su Y, Yang M et al: Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol Cancer 2019, 18(1): 95.
- Yan D, Dong W, He Q, Yang M, Huang L, Kong J, Qin H, Lin T, Huang J: Circular RNA circPICALM sponges miR-1265 to inhibit bladder cancer metastasis and influence FAK phosphorylation. EBioMedicine 2019, 48: 316-331.
- Bi J, Liu H, Dong W, Xie W, He Q, Cai Z, Huang J, Lin T: Circular RNA circ-ZKSCAN1 inhibits bladder cancer progression through miR-1178-3p/p21 axis and acts as a prognostic factor of recurrence. Mol Cancer 2019, 18(1): 133.
- Li Y, Wan B, Liu L, Zhou L, Zeng Q: Circular RNA circMTO1 suppresses bladder cancer metastasis by sponging miR-221 and inhibiting epithelial-to-mesenchymal transition. Biochem Biophys Res Commun 2019, 508(4): 991-996.
- Zheng F, Wang M, Li Y, Huang C, Tao D, Xie F, Zhang H, Sun J, Zhang C, Gu C et al: CircNR3C1 inhibits proliferation of bladder cancer cells by sponging miR-27a-3p and downregulating cyclin D1 expression. Cancer Lett 2019, 460: 139-151.
- Yu XY, Ma CQ, Sheng YH: circRNA CRIM1 regulates the migration and invasion of bladder cancer by targeting miR182/Foxo3a axis. Clin Transl Oncol 2022, 24(6): 1195-1203.
- Yang D, Qian H, Fang Z, Xu A, Zhao S, Liu B, Li D: Silencing circular RNA VANGL1 inhibits progression of bladder cancer by regulating miR-1184/IGFBP2 axis. Cancer Med 2020, 9(2): 700-710.
- Yang C, Wu S, Wu X, Zhou X, Jin S, Jiang H: Silencing circular RNA UVRAG inhibits bladder cancer growth and metastasis by targeting the microRNA-223/fibroblast growth factor receptor 2 axis. Cancer Sci 2019, 110(1): 99-106.
- Zhang H, Shen Y, Li Z, Ruan Y, Li T, Xiao B, Sun W: The biogenesis and biological functions of circular RNAs and their molecular diagnostic values in cancers. J Clin Lab Anal 2020, 34(1): e23049.
- Bi J, Liu H, Cai Z, Dong W, Jiang N, Yang M, Huang J, Lin T: Circ-BPTF promotes bladder cancer progression and recurrence through the miR-31-5p/RAB27A axis. Aging (Albany NY) 2018, 10(8): 1964-1976.
- Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ et al: PRMT5 Circular RNA Promotes Metastasis of Urothelial Carcinoma of the Bladder through Sponging miR-30c to Induce Epithelial-Mesenchymal Transition. Clin Cancer Res 2018, 24(24): 6319-6330.
- Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, Chen J: Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett 2017, 403: 305-317.
- Wu Z, Huang W, Wang X, Wang T, Chen Y, Chen B, Liu R, Bai P, Xing J: Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol Med 2018, 24(1): 40.
- Sun M, Zhao W, Chen Z, Li M, Li S, Wu B, Bu R: Circular RNA CEP128 promotes bladder cancer progression by regulating Mir-145-5p/Myd88 via MAPK signaling pathway. Int J Cancer 2019, 145(8): 2170-2181.
- Su Y, Feng W, Shi J, Chen L, Huang J, Lin T: circRIP2 accelerates bladder cancer progression via miR-1305/Tgf-beta2/smad3 pathway. Mol Cancer 2020, 19(1): 23.
- Zhang X, Liu X, Jing Z, Bi J, Li Z, Liu X, Li J, Li Z, Zhang Z, Kong C: The circINTS4/miR-146b/CARMA3 axis promotes tumorigenesis in bladder cancer. Cancer Gene Ther 2020, 27(3-4): 189-202.
- Yao J, Qian K, Chen C, Liu X, Yu D, Yan X, Liu T, Li S: ZNF139/circZNF139 promotes cell proliferation, migration and invasion via activation of PI3K/AKT pathway in bladder cancer. Aging (Albany NY) 2020, 12(10): 9915-9934.
- Ma X, Ying Y, Sun J, Xie H, Li J, He L, Wang W, Chen S, Shen H, Yi J et al: circKDM4C enhances bladder cancer invasion and metastasis through miR-200bc-3p/ZEB1 axis. Cell Death Discov 2021, 7(1): 365.
- Zhu J, Luo Y, Zhao Y, Kong Y, Zheng H, Li Y, Gao B, Ai L, Huang H, Huang J et al: circEHBP1 promotes lymphangiogenesis and lymphatic metastasis of bladder cancer via miR-130a-3p/TGFbetaR1/VEGF-D signaling. Mol Ther 2021, 29(5): 1838-1852.
- Feng D, Lv J, Li K, Cao Q, Han J, Yu H, Cheng Y, Zhuang J, Cai L, Yang H et al: CircZNF609 promotes bladder cancer progression and inhibits cisplatin sensitivity via miR-1200/CDC25B pathway. Cell Biol Toxicol 2022, https://doi.org/10.1007/s10565-022-09715-3. Epub ahead of print.
- Yang J, Qi F, Tan B, Dai G, Chen R, Wan W, Cheng B, Xue B: circSPECC1 promotes bladder cancer progression via regulating miR-136-5p/GNAS axis. Pathol Res Pract 2022, 234: 153914.
- Yang J, Liu X, Dai G, Qu L, Tan B, Zhu B, Qi F, Gai X, Cheng B: CircNT5E promotes the proliferation and migration of bladder cancer via sponging miR-502-5p. J Cancer 2021, 12(8): 2430-2439.
Annals of urologic oncology
p-ISSN: 2617-7765, e-ISSN: 2617-7773
 Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript