Research Article | Open Access
Long Non-coding RNA INE1 Induced Autophagy Promotes Sensitivity of Prostate Cancer Cells to Cisplatin
Hezhen Chu1, Kongdong Li2, Jie Gu2, Wenchao Xie3, Yimin Xie3 , Jun Ma1
1Department of Urology, Yixing Traditional Chinese Medicine Hospital, Yixing, Jiangsu, China.
2School of Life Sciences, Jiangsu University, Zhenjiang, Jiangsu, China.
3Department of Urology, Affiliated Hospital of Jiangsu University-Yixing Hospital, Yixing, Jiangsu, China.
Correspondence: Jun Ma (Department of Urology, Yixing Traditional Chinese Medicine Hospital, Yixing, 214200, China; Email: yxzyymajun@126.com).
Annals of Urologic Oncology 2022; 5(2): 89-100. https://doi.org/10.32948/auo.2022.11.24
Received: 16 Nov 2022 | Accepted: 21 Nov 2022 | Published online: 29 Nov 2022
Key words prostate cancer, LncRNA, INE1, autophagy, apoptosis, cisplatin
Long non-coding RNA (lncRNA) has been documented in TCGA database and has enormous potential in cancer research. It is a group of non-protein coding transcripts with ≥200 nucleotides, widely expressed (about 60,000 lncRNA) in the human genome and widespread both in nucleus and cytoplasm [6, 7]. In the past three decades, more than hundreds of cancer associated lncRNAs were identified and have been reported to play an important role in regulating cancer development [6, 8, 9], including prostate cancer [10, 11]. However, the functions of lncRNAs in prostate cancer initiation, progression, and their clinical utility in disease management are still infancy [12]. The lncRNA inactivation escape 1 (INE1) also known as non-protein coding RNA 10 is located on the human chromosome Xp11.3 and consists of 1014 bases and contains one exon. As an X chromosome gene, INE1 is intronless and was identified because its transcription escapes X inactivation in females [13]. Through a microarray analysis and an integrated functional gene map, lncRNA INE1 was identified as playing possible roles in pathogenesis of cerebral aneurysms and neurogenetic disorders [14, 15]. Recently, lncRNA INE1 was documented to play a critical role in the regulation of prostate cancer autophagy [16]. However, the function and mechanism of lncRNA INE1 in prostate cancer has not yet been elucidated.
The serine/arginine-rich splicing factors (SRSFs) belong to the serine arginine-rich protein family, which plays an important role in the recruit and assemble spliceosome to regulate the splicing process of precursor RNA. In tumors, SRSFs act as oncoproteins in disrupting both DNA stability and normal protein expression pattern, contributing to proliferation, migration and apoptosis resistance of tumor cells [17]. In prostatic cancer patients, SRSFs such as SRSF2, SRSF5, SRSF7 and SRSF8 were identified as negative prognostic biomarkers, due to their higher expression that correlated with poor overall survival [18]. The regulation of SRSFs by lncRNAs were shown to be involved in the progression of tumors. LncRNA PANDAR dictates the chemoresistance of ovarian cancer via regulating SFRS2-mediated p53 phosphorylation [19]. LncRNA LINC02580 suppresses the invasion of hepatocellular carcinoma by targeting SRSF1 [20]. SRSF1 splicing in gastric cancer was enhanced by lncRNA FENDRR [21]. However, the role of lncRNA on SRSFs in tumor progression still needs further evaluation.
In the present study, we identified the correlation of lncRNA INE1 with the survival times of patients with prostate cancer and the clinico-pathological features. In order to explore the role of INE1 in prostate cancer, we detected the cell viability, cell proliferation, autophagy and apoptosis in siRNA INE1 knockdown or cisplatin treated prostate cancer cells. Moreover, the role of lncRNA INE1 on splicing factor SRSF2 was detected in prostate cancer cells.
MTT (3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide) was purchased from Solarbio (Beijing, China). The anti-SRSF2, Caspase-9, Caspase-3 and β-actin antibody were purchased from Cell Signaling Technology (Shanghai, China). The anti-Ki-67 antibody, Alexa Fluor 488 goat anti-mouse IgG and DAPI were purchased from Thermo Fisher Scientific (Shanghai, China). The terminal desoxynucleotidyl transferase (dUTP) nick end labeling (TUNEL) kit was purchased from Beyotime (Shanghai, China). RNA-to-cDNA kit and SYBR Green I kit were purchased from Vazyme (Nanjing, China). Mediums, FBS, penicillin, and streptomycin were purchased from Gibco (CA, USA). PVDF membranes and chemiluminescent reagent were purchased from Merck (Shanghai, China). Cisplatin (CDDP) was purchased from Master of Bioactive Molecules (Shanghai, China), Chloroquine (CQ) was purchased from Sigma (USA).
Cell culture and treatments
Prostate cancer cell lines DU145, LNCaP and PC-3, and normal human prostate stromal cell WPMY-1 were obtained from American Tissue Culture Collection (ATCC, Manassas, VA, USA). DU145 cells were cultured in MEM; LNCaP cells were cultured in RPMI 1640 (pH 7.4) medium; PC-3 cells were cultured in Ham’sF12k (pH 7.4); WPMY-1 cells were cultured in DMEM medium (pH 7.4), and supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, and 100 mg/ml streptomycin (pH 7.4) in a humidified atmosphere containing 5 % CO2. For some experiments, the cells were treated with cisplatin (2, 5, 10 μM) for 24 h.
Cell migration detection
Cell migratory capacity was assessed using the wound-healing assay. Cells were seeded in 6-wells plate (2 x 105 cells/well) in 2 mL complete medium. Once reaching 90-95% confluence, a scratch was created manually through the confluent monolayer using a white tip (2-20 μL). The culture medium was then aspirated, and wells were washed with PBS to remove cellular debris. Fresh medium without FBS was then added along with the indicated treatments. Images were taken at baseline (0 h) and at the indicated time points by Olympus IX73 microscopy (Japan). Microscope software was used to measure the width of the scratch, which reflected the extent of cell migration. The distance migrated was measured in μm.
MTT assays
DU145 and PC-3 cells were seeded in 6-well and transfected with vector control, INE1 siRNA-2 and siRNA-3 for 24 hrs, cells were collected and seeded in 96-well with a density of 2 × 103 cells/well. In some experiments, the cells were treated with CDPP (2 μM) for 24 h with or without pretreatment of CQ (20 μM) 1 h. After culture for 24 hrs, the cell viability was measured by the MTT assay. In brief, cells were incubated in 100 μL MTT solution (0.5 mg/ml in MEM medium) in 96-well plate for 4 h before the end of incubation. The supernatant was then discarded, and 100 μL DMSO was added to dissolve the colored product. The absorbance was measured at 540 nm (690 nm as reference) using a Synergy H4 Hybrid Multi-Mode Microplate Reader (BioTek, USA).
Real-time quantitative PCR (RT-qPCR) analysis
Cell total RNA was isolated by using Trizol (Life Technologies, MD, USA). All RNA isolations were performed as directed by the manufacturer. Gel electrophoresis and staining confirmed the purity and integrity of the samples. Quantification of RNA was based on spectrophotometric analysis at 260/280 nm. cDNA was generated from 10 μg total RNA using a RNA-to-cDNA kit. Real-time PCRs were carried out by ABI7300 system using SYBR Green I kit. Gene-specific primers for INE1-F: GGAAGGCCTGAGTTCTGCAA, INE1-R: CCCACCTCAGGATCTTTGCG, and β-Actin-F: CTCCATCCTGGCCTCGECTGT, β-Actin-R: GCTGTCACCTTCACCGTTCC were used. The occurrence of primer-dimers and secondary products was inspected using melting curve analysis and agarose gel electrophoresis. Control amplification was done either without reverse transcriptase or without RNA. The relative expression ratio of the target gene was calculated according to their threshold cycle Ct values.
Knockdown of INE1 and SRSF2
DU145 and PC-3 cells were seeded in 6-well with a density of 2 × 105 cells/well and cultured overnight. Then the cells were transfected with INE1 siRNAs packaged by Lipo2000 reagent according to manufacturer instructions (Thermo Fisher Scientific, WI, USA). At 24 hrs after transfection the cells were harvested for further evaluation. The siRNAs used in this study were INE1 siRNA-1: sense 5?- CGUGAGGUAUAUAGUUAAUTT-3? and antisense 5?- AUUAACUAUAUACC UCACGTT-3?; INE1 siRNA-2: sense 5?-GGAGAAUGGUUUGUAGUAUTT-3? and antisense 5?-AUACUACAAACCAUUCUCCTT-3?; INE1 siRNA-3: sense 5?-GUCC ACAGCUUCCCUUUAUTT-3?; and antisense 5?-AUAAAGGGAAGCUGUGGACT T-3?; SRSF2 siRNA-1: sense 5?-GCGGUGUCCUCUUAAGAAATT-3? and antisense 5?-UUUCUUAAGAGGACACCGCTT -3?; SRSF2 siRNA-2: sense 5?-GAGCAGGU UUGUCUUUAAATT -3? and antisense 5?-UUUAAAGACAAACCUGCUCTT-3?; and siRNA control: sense 5?-UUCUCCGAACGUGUCACGUTT-3?, antisense 5?-ACGUGACACGUUCGGAGAATT-3? from GenePharm (Shanghai, China).
Western blot analysis
The cells were lysed in radioimmunoprecipitation assay (RIPA) buffer. After centrifugation at 13,000 g for 15 min at 4 oC, the supernatant was collected and the total protein concentration was determined by BCA Assay (Protein Quantification Kit) (Thermo Fisher Scientific, USA). The protein lysates containing 40 μg total cellular protein in RIPA buffer were subjected to electrophoresis on 10% polyacrylamide gels. The gels were then blotted onto PVDF membranes. Western blotting was conducted using rabbit monoclonal antibodies against LC3B, P62, Caspase-9, Caspase-3, SRSF2, and β-actin antibody (1:1000), followed by incubation with horseradish peroxidase-conjugated goat anti-rabbit antibody (1:4000). Specific bands were visualized using chemiluminescent reagent, and the images were taken by Tanon-5200 Multi Automatic Chemiluminescence Image Analysis System. The relative protein levels were analyzed by Image J.
Immunofluorescence and fluorometric cell death detection
DU145 and PC-3 cells were seeded in 6-well and transfected with siRNA control, INE1 siRNA-2 and siRNA-3 for 24 hrs, with or without treatment of cisplatin (2 μM) for 24 hours in apoptosis detection experiments. Then the cells were fixed for 30 min in 4% Formaldehyde (FA, Sigma-Aldrich) and permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) in PBS for 20 min. After blocking with 3% normal goat serum, the cells were incubated with mouse anti-Ki-67 (1:100) antibody overnight at 4 oC, followed by 1 h incubation with Alexa Fluor 488 goat anti-mouse IgG (1:200,). After washing twice with PBS, cell nucleus was stained by the DAPI for several minutes. The cells were washed in PBS for 10 min and the cells were mounted, then examined by Olympus IX73 microscopy (Japan). Fluorometric cell death was detected by TUNEL assay according to the manufacturer’s instructions, and counterstained with DAPI as previously described. Then the images were taken by Olympus IX73 microscopy (Japan).
Statistical analysis of data
Prostate cancer dataset (TCGA) (https://www.genenames. org/) was searched where 2708 lncRNAs (http://www.genenames.org) and 2687 lncRNAs (http://www. cbioportal.org) were analyzed by two different software. We selected all 499 tumors samples (498 patients) identified in the primary search. The primary search parameters included mutations, copy number alterations (CNA) from GISTIC and mRNA expression (RNA seq data) with the default setting. For the secondary search, we focused on RNA seq data and analyzed the correlation between their expressions and the survival time of prostate cancer patients. All available lncRNAs were sorted by alteration frequency at the cBioPortal. Those lncRNAs with significant log-rank p values were entered in the candidate pool to be considered for further selection. P values less than 5% were determined as significant. Association with the INE1 expression levels and clinic-pathological features were analyzed with the chi-square (χ2) test for categorical variables and the Wilcoxon test for continuous variables. The statistical analyses were performed using the SPSS 20.0 software analysis. The student t-test was used to compare the results and expressed as mean ± S.D. between control and treatment groups. P < 0.05 was considered statistically significant.
Analysis revealed that the lncRNA INE1 is the top one related to both overall survival and disease free survival time (Figure 1A, B). According to the GEPIA analysis, the disease free survival time and overall survival time of patients with higher INE1 expression were significantly shorter than those with lower INE1 expression (P < 0.05) (Figure 1C, D). As shown in Table 1, there was no significant statistical correlation between age, lymph node metastasis and count, PSA results with the expression of INE1 (P > 0.05), but it positively correlated with tumor stage, biochemical recurrence, disease recurrence and Gleason pattern (P < 0.05). It suggested that lncRNA INE1 might act as an oncogene promoting tumor malignancy and recurrence.
Higher INE1 expression in prostate cancer cells promotes cell viability
To identify the functions of INE1, firstly, we compared the mRNA expression of INE1 in prostate cell lines DU145, LNCaP and PC-3 with normal human prostate stromal cell WPMY-1 cells. It showed that INE1 expression in prostate cancer cell lines was significantly higher than in WPMY-1 cells (Figure 2A). To further clarify the roles of INE1 in prostate cancer growth, small interfering RNA was applied. The results of RT-PCR showed the second and third pair of siRNA had the higher efficiency on INE1 silencing in DU145 and PC-3 cells (Figure 2B). Furthermore, MTT assays indicated that siRNA silencing INE1 for 24 h resulted in significant inhibition of cell viability in both DU145 and PC-3 cells (Figure 2C). However, the immunofluorescence staining of cell proliferation biomarker Ki-67 showed that there was no difference of the expression of Ki-67 in INE1 knockdown cells compared to negative control in both cell lines (Figure 2D).
Silencing INE1 expression induces autophagy and pro-apoptosis of prostate cancer cells
To further clarify INE1 promote cell viability, the expressions of the biomarkers of cell autophagy and apoptosis were detected by western blotting. Knockdown INE1 by siRNAs increased the expression of early autophagy inducer LC3-II but did not change the expression of p62 in both DU145 and PC-3 cells (Figure 3A-C). In addition, INE1-siRNAs increased the expression of caspase-9 and caspase-3 in both DU145 and PC-3 cells (Figure 3A, D, E). These results indicated that silencing INE1 expression might induce autophagy signal and pro-apoptosis in prostate cancer cells.
Silencing INE1 promotes sensitivity of prostate cancer cells to cisplatin
To further identify lncRNA INE1-regulated autophagy in prostate cancer, the cells were treated with cisplatin and/or autophagy inhibitor CQ. The results of MTT assay showed that low dose of cisplatin (2 μM) inhibited cell viability of prostate cancer cells, which were augmented by autophagy inhibitor CQ (Figure 4A, B). While the results of Western blotting showed that low dose of cisplatin treatment significantly suppressed the expression of LC3-II, but did not significantly change the expression of p62, caspase-9 and caspase-3 in both DU145 and PC-3 cells (Figure 4C-E). The result of qRT-PCR showed that cisplatin induced the expression of INE1 in DU145 cells, and high dose of cisplatin induced its expression in PC-3 cells (Figure 4F). In addition, the result of TUNEL staining showed that INE1-siRNAs increased low dose of cisplatin treatment -induced nuclear DNA damage in both DU145 and PC-3 cells (Figure 4G). These results suggest that silencing lncRNA INE1-induced autophagy and apoptosis aggravated cisplatin-inhibited cell viability in prostate cancer.
INE1 plays an anti-apoptotic role by regulating the expression of the splicing factor SRSF2
To identify the anti-apoptotic role INE1 in prostate cancer cells, we detected the regulation of INE1 on SRSF2, which is well known splicing factor and plays anti-apoptotic role in cancer cells [22, 23]. The result of Western blotting showed that knockdown INE1 by siRNAs decreased the protein level of SRSF2 in both DU145 and PC-3 cells (Figure 5A, B). In addition, the results of Western blotting and TUNEL staining showed that knockdown SRSF2 by siRNA increased the expressions of apoptosis markers Caspase-9 and Caspase-3 and induce DNA damage (Figure 5C-G). These results suggest that INE1 play an anti-apoptotic role by targeting the expression of the splicing factor SRSF2 in prostate cancer cells.
|
Table 1. Correlation analysis of INE1 expression level with clinicopathological features of patients with prostate cancer. |
||||
|
Variable |
Total(%) |
Expression of INE1 level |
P value |
|
|
Low |
High |
|||
|
Age |
n=498 |
|
|
0.520 |
|
≦60 |
223 (44.8%) |
210 |
13 |
|
|
>60 |
275 (55.2%) |
255 |
20 |
|
|
Tumor Stage Code (AJCC) |
n=491 |
|
0.003* |
|
|
T1 |
0 |
0 |
0 |
|
|
T2 |
187 (38.1%) |
181 |
6 |
|
|
T3 |
293 (59.7%) |
271 |
22 |
|
|
T4 |
11 (2.2%) |
8 |
3 |
|
|
Biochemical recurrence index |
n=430 |
|
0.033* |
|
|
No |
371 (86.2%) |
349 |
22 |
|
|
Yes |
59 (13.7%) |
51 |
30 |
|
|
Disease recurrence |
n=492 |
|
|
0.045* |
|
Disease Free |
400 (81.3%) |
379 |
21 |
|
|
Recurred/Progressed |
92 (18.7%) |
82 |
10 |
|
|
Lymph node metastasis (N) |
n=406 |
|
|
0.705 |
|
N0 |
325 (80.0%) |
301 |
24 |
|
|
N1 or above |
81 (20.0%) |
76 |
5 |
|
|
Gleason score |
n=498 |
|
|
0.014* |
|
Gleason ≦7 |
210 (42.2%) |
200 |
10 |
|
|
Gleason >7 |
288 (57.8%) |
265 |
23 |
|
|
PSA result |
n=440 |
410 |
30 |
0.385 |
|
Lymph node count |
n=423 |
391 |
32 |
0.873 |
|
Tumor Stage Code: American Joint Committee on Cancer Tumor Stage Code; Lymph Node metastasis (N): Positive Finding Lymph Node Hematoxylin and Eosin Staining Microscopy Count |
||||
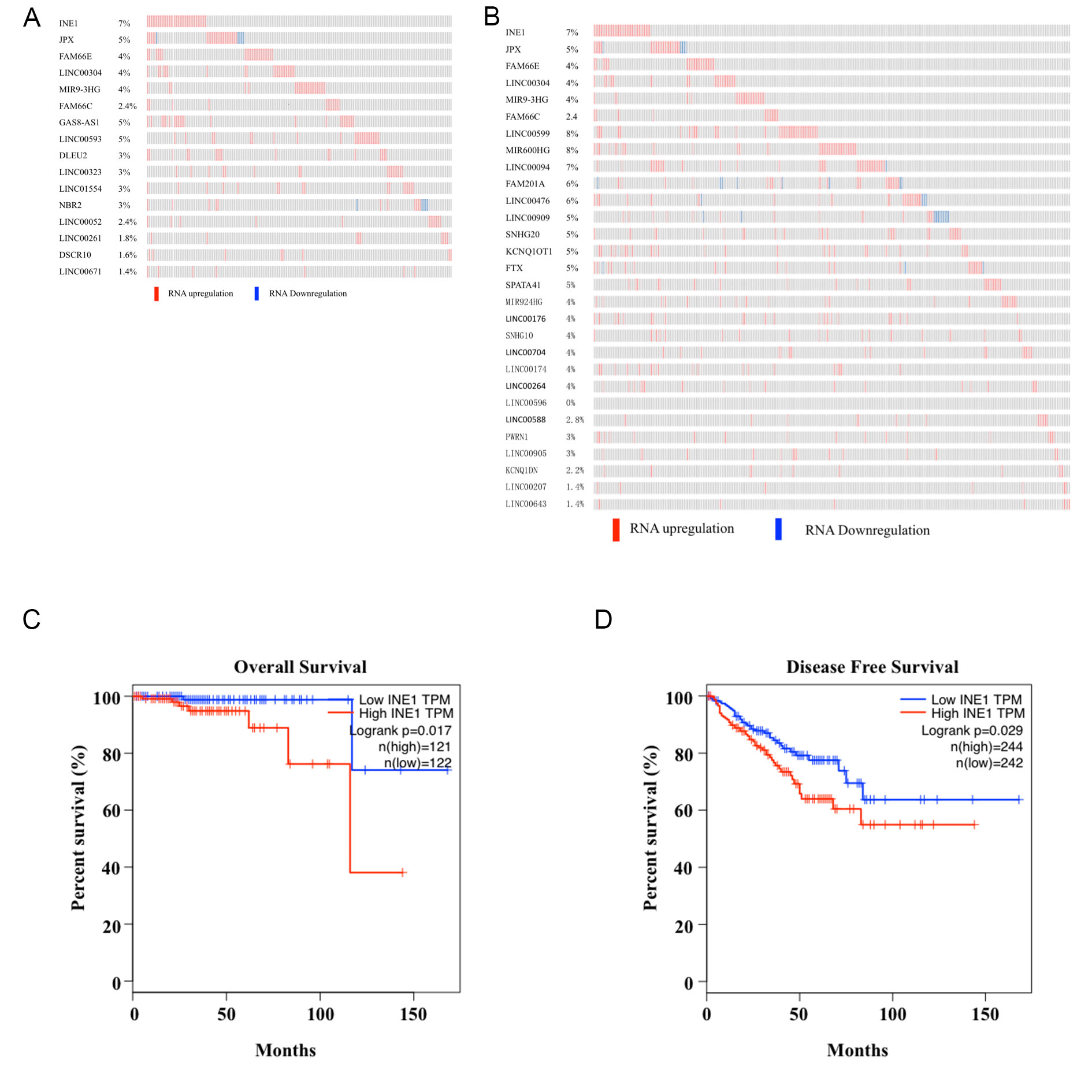 Figure 1. Signature of INE1 gene expression with survival time of the prostate cancer patients. Signature of LncRNAs for Overall Survival (A) and Disease Free Survival (B) based on mRNA expression. GEPIA analysis shows Kaplan-Meier survival curves of Overall Survival (C) and Disease Free Survival (D) for prostate cancer patients compared high INE1 expression with low expression.
Figure 1. Signature of INE1 gene expression with survival time of the prostate cancer patients. Signature of LncRNAs for Overall Survival (A) and Disease Free Survival (B) based on mRNA expression. GEPIA analysis shows Kaplan-Meier survival curves of Overall Survival (C) and Disease Free Survival (D) for prostate cancer patients compared high INE1 expression with low expression.
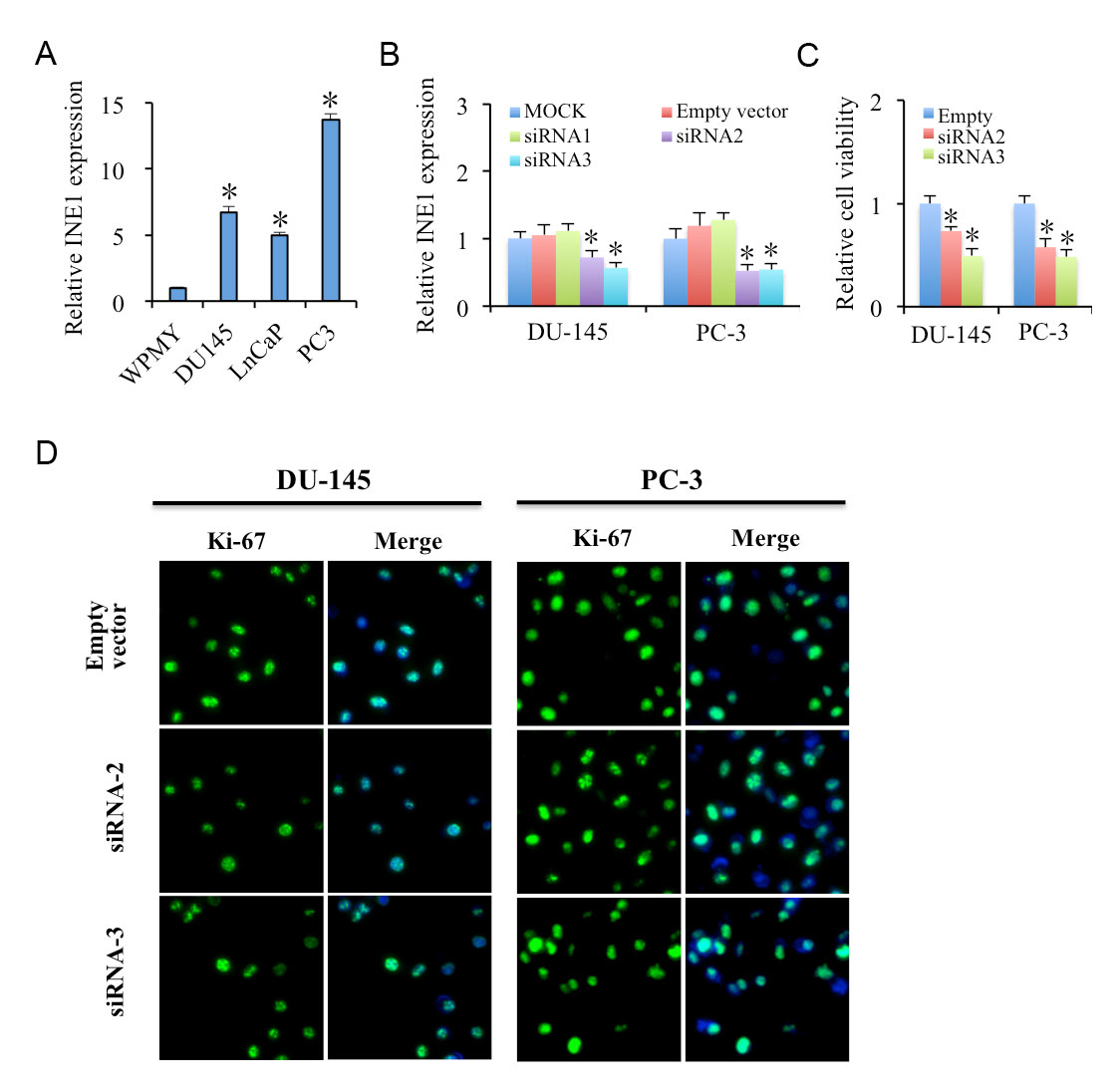 Figure 2. Knockdown INE1 inhibits cell viability in prostate cancer cells. (A) INE1 is highly expressed in prostate cancer cell lines DU145, LNCaP and PC-3 as compared to normal human prostate stromal cell WPMY-1 cells, determined by RT-PCR. Results are presented as mean ± SD (n=3). * P<0.05, using Student’s t-test. (B) Expression of INE1 in knockdown DU-145 and PC-3 cells as compared to siRNA control. (C) INE1 knockdown in DU-145 and PC-3 cells suppresses cell growth as detected by MTT assays. Results are presented as mean ± SD (n=4). * Statistical significance between control and siRNA treatments, * P<0.05, using Student’s t-test. (D) INE1 knockdown dose not affect cell proliferation in DU-145 and PC-3 cells as detected by Ki-67 immunofluorescence. Ki-67: Green, DAPI: Blue. Scalar bar: 50 μm.
Figure 2. Knockdown INE1 inhibits cell viability in prostate cancer cells. (A) INE1 is highly expressed in prostate cancer cell lines DU145, LNCaP and PC-3 as compared to normal human prostate stromal cell WPMY-1 cells, determined by RT-PCR. Results are presented as mean ± SD (n=3). * P<0.05, using Student’s t-test. (B) Expression of INE1 in knockdown DU-145 and PC-3 cells as compared to siRNA control. (C) INE1 knockdown in DU-145 and PC-3 cells suppresses cell growth as detected by MTT assays. Results are presented as mean ± SD (n=4). * Statistical significance between control and siRNA treatments, * P<0.05, using Student’s t-test. (D) INE1 knockdown dose not affect cell proliferation in DU-145 and PC-3 cells as detected by Ki-67 immunofluorescence. Ki-67: Green, DAPI: Blue. Scalar bar: 50 μm.
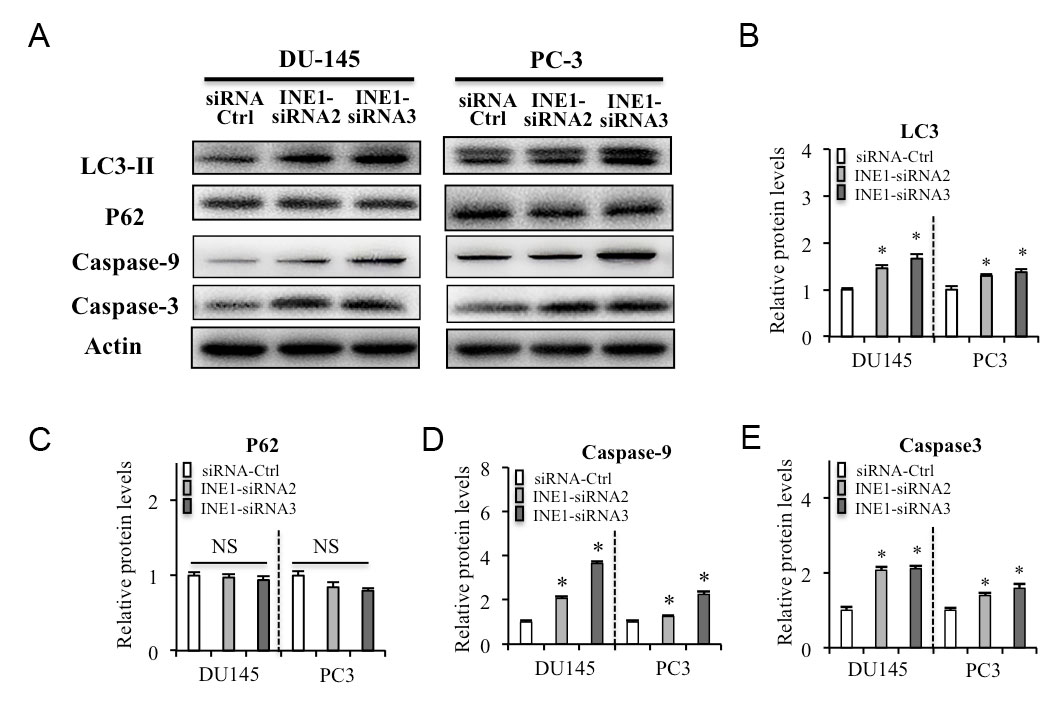 Figure 3. Silencing INE1 on autophagy and pro-apoptosis in prostate cancer cells. (A) Effects of INE1-siRNAs on the expressions of the autophagy biomarkers LC3-II, P62, and pro-apoptosis biomarkers Caspase-9 and Caspase-3 in DU-145 and PC-3 cells detected by western blotting. (B-E) Relative protein levels were analyzed by Image J. NS: no significant difference. *P<0.05, Statistical significance between indicated groups, using Student’s t-test.
Figure 3. Silencing INE1 on autophagy and pro-apoptosis in prostate cancer cells. (A) Effects of INE1-siRNAs on the expressions of the autophagy biomarkers LC3-II, P62, and pro-apoptosis biomarkers Caspase-9 and Caspase-3 in DU-145 and PC-3 cells detected by western blotting. (B-E) Relative protein levels were analyzed by Image J. NS: no significant difference. *P<0.05, Statistical significance between indicated groups, using Student’s t-test.
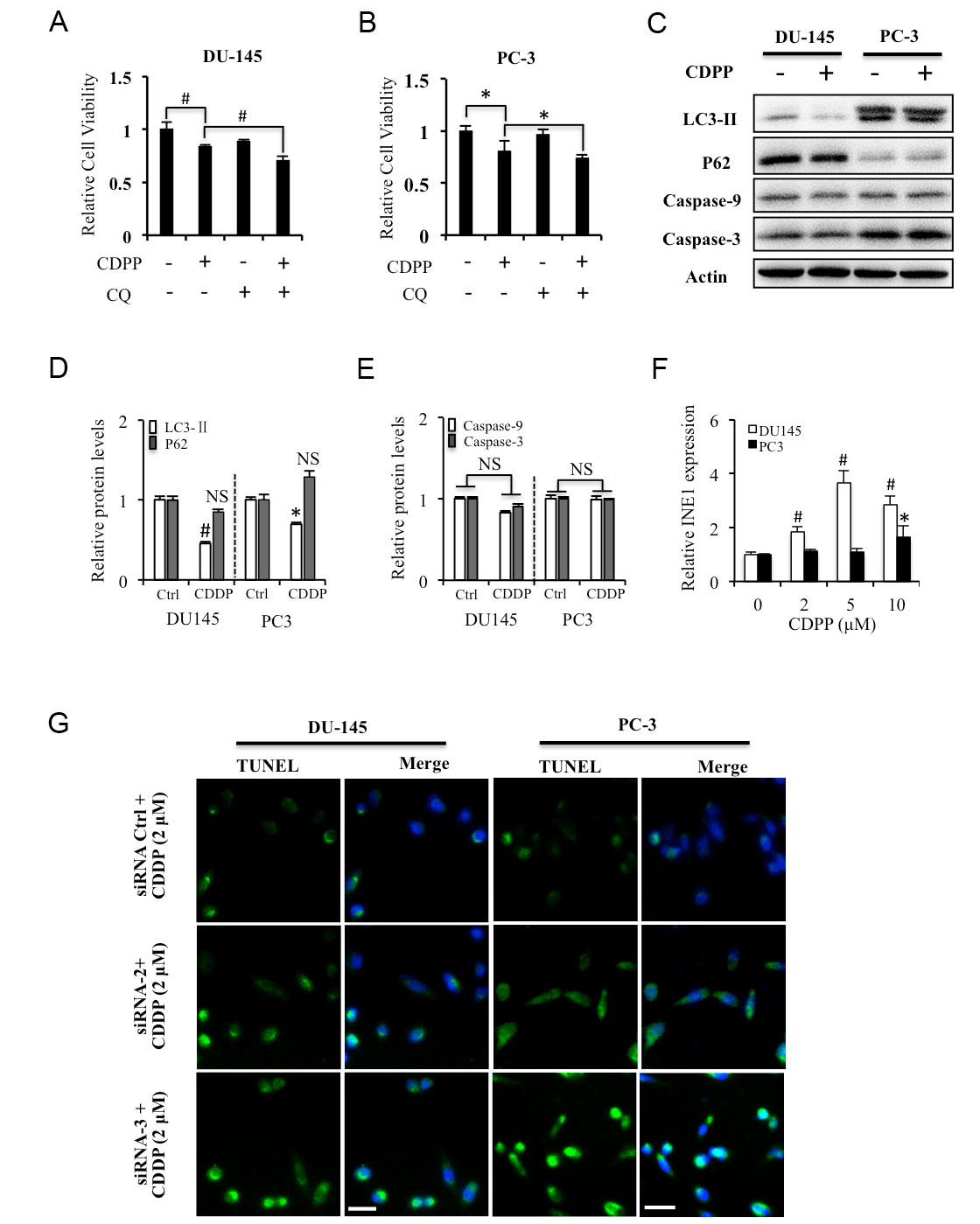 Figure 4. Silencing INE1 on cisplatin induced apoptosis in prostate cancer cells. (A, B) Effects of CDPP and CQ on the cell viability in DU-145 and PC-3 cells detected by MTT assay. (C) Effects of CDPP on the expressions of the autophagy biomarkers LC3-II, P62, and pro-apoptosis biomarkers Caspase-9 and Caspase-3 in DU-145 and PC-3 cells detected by western blotting. (D, E) Relative protein levels were analyzed by Image J. (F) Effects of CDPP on the expressions of INE1 expression detected by RT-qPCR. (G) Effects of CDPP on DNA damage in INE1 knockdown DU-145 and PC-3 cells detected by TUNEL staining. Green: TUNEL, Blue: DAPI. Scalar bar: 50 μm. CDPP: cisplatin, 2 μM. CQ: Chloroquine, 20 μM. NS: no significant difference. *P<0.05, # P<0.01, Statistical significance between indicated groups, using Student’s t-test.
Figure 4. Silencing INE1 on cisplatin induced apoptosis in prostate cancer cells. (A, B) Effects of CDPP and CQ on the cell viability in DU-145 and PC-3 cells detected by MTT assay. (C) Effects of CDPP on the expressions of the autophagy biomarkers LC3-II, P62, and pro-apoptosis biomarkers Caspase-9 and Caspase-3 in DU-145 and PC-3 cells detected by western blotting. (D, E) Relative protein levels were analyzed by Image J. (F) Effects of CDPP on the expressions of INE1 expression detected by RT-qPCR. (G) Effects of CDPP on DNA damage in INE1 knockdown DU-145 and PC-3 cells detected by TUNEL staining. Green: TUNEL, Blue: DAPI. Scalar bar: 50 μm. CDPP: cisplatin, 2 μM. CQ: Chloroquine, 20 μM. NS: no significant difference. *P<0.05, # P<0.01, Statistical significance between indicated groups, using Student’s t-test.
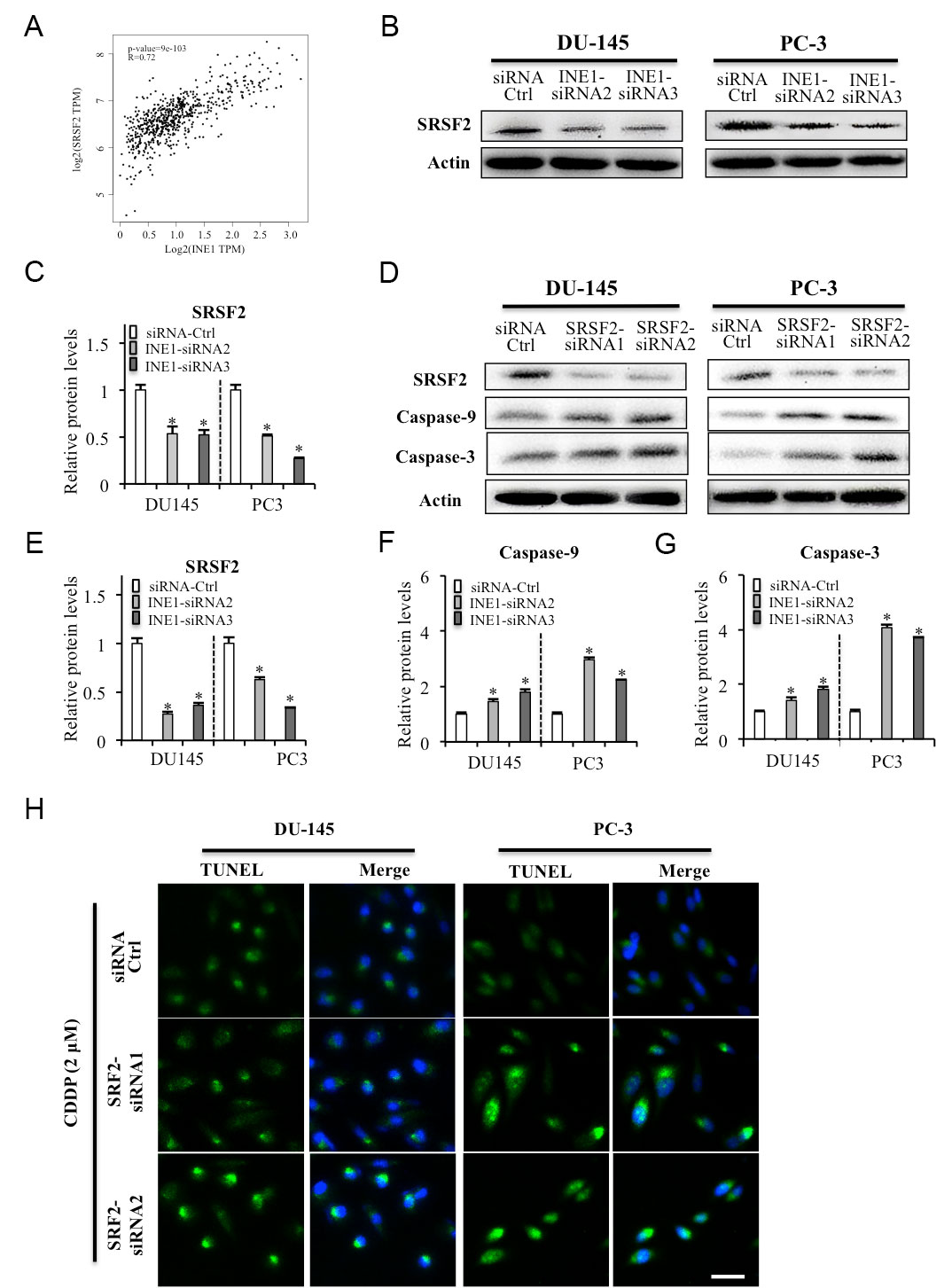 Figure 5. Roles of INE1 targeting SRSF2 on cell apoptosis in prostate cancer cells. (A) Correlation of INE1 with SRSF2. (B, C) Expression of SRSF2 in INE1 knockdown DU145 and PC-3 cells detected by western blotting. (D-G) Efficacies of SRSF2 siRNAs on the expression of apoptosis marker Caspase-9 and Caspase-3 as detected by western blotting. β-Actin was used as loading reference. Relative protein levels were analyzed by Image J. *P<0.05, Statistical significance between indicated groups, using Student’s t-test. (H) CDDP induced cell apoptosis in silencing SRSF2 DU145 and PC-3 cells detected by TUNEL staining. CDDP: cisplatin, 2 μM. Green: TUNEL, Blue: DAPI. Scalar bar: 50 μm.
Figure 5. Roles of INE1 targeting SRSF2 on cell apoptosis in prostate cancer cells. (A) Correlation of INE1 with SRSF2. (B, C) Expression of SRSF2 in INE1 knockdown DU145 and PC-3 cells detected by western blotting. (D-G) Efficacies of SRSF2 siRNAs on the expression of apoptosis marker Caspase-9 and Caspase-3 as detected by western blotting. β-Actin was used as loading reference. Relative protein levels were analyzed by Image J. *P<0.05, Statistical significance between indicated groups, using Student’s t-test. (H) CDDP induced cell apoptosis in silencing SRSF2 DU145 and PC-3 cells detected by TUNEL staining. CDDP: cisplatin, 2 μM. Green: TUNEL, Blue: DAPI. Scalar bar: 50 μm.
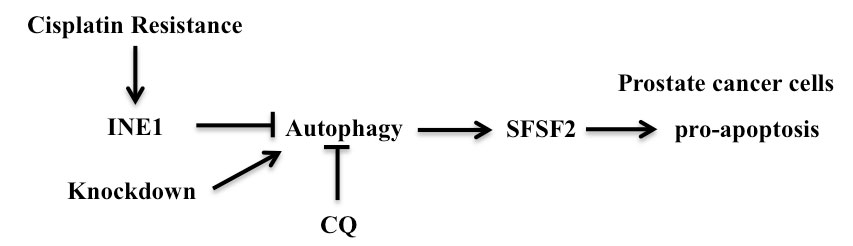 Figure 6. Schematic diagram to show the mechanism that relationship between LncRNA FAM66C and prostate cancer.
Figure 6. Schematic diagram to show the mechanism that relationship between LncRNA FAM66C and prostate cancer.
Previous studies have explored that lncRNAs enhances drug resistance, promotes ferroptosis and bone metastasis in prostate cancer [25-28], as well as promote proliferation, migration, and invasion in prostate cancer cells [29]. It is also reported that lncRNAs regulates autophagy in prostate cancer cells. Upregulation of LncRNA IDH1-AS1 promotes tumor growth in prostate cancer by regulating ATG5-mediated autophagy [30]. Silencing lncRNA PRRT3-AS1 inhibits prostate cancer cell proliferation and promotes apoptosis and autophagy [31]. LncRNA BCAR4 enhances autophagy in prostate cancer cells [32]. Autophagy is a process that delivers cellular material to lysosomes for degrading, recycling, and providing energy and molecular precursors. It suggests that promoting autophagy might prevent cancer development in premalignant lesions, but both enhancing and inhibiting autophagy could be therapeutic strategies in advanced cancers [33]. INE1 was indicated as an autophagy-associated lncRNA in prostate cancer [16]. Our results suggest that silencing INE1 expression induce autophagy signal associated with pro-apoptosis in prostate cancer cells.
Cisplatin (CDDP) is a widely used chemotherapy agent for various types of cancers. It can also induce autophagy in cancer cells, including prostate cancer cells at high dose (5 μM). Low dose of CDDP suppressed the expression of autophagy-related molecules including LC3-II, P62, ATG5, and ATG7 in rat astrocytes in vitro [34]. Our results showed that low dose of CDDP (2 μM) reduced the expression of LC3-II, but did not change the expression of P62, as well as the pro-apoptosis biomarkers caspase-9 and caspase-3. It suggested that low dose of CDDP inhibited the early autophagy signal and could not induce cell apoptosis in prostate cancer cells. The regulation of autophagy could modulate the sensitivity of cancer cells to cisplatin. Ambra1 induces autophagy and desensitizes prostate cancer cells (DU145) to CDDP (10 μM) [35]. Knockdown of cystic fibrosis transmembrane conductance regulator (CFTR) enhances sensitivity of prostate cancer cells (LNCaP/CP) to CDDP (40 μM) via inhibition of autophagy [36]. It is also reported miR-205 impaired the autophagic flux and enhanced CDDP (5, 10 μM) cytotoxicity in androgen-refractory prostate cancer cells (DU145 and PC-3) [37]. Circular RNA circCUL2 regulate cisplatin sensitivity through miR-142-3p/ROCK2-mediated autophagy activation in gastric cancer cells [38]. Artesunate impairs growth in cisplatin-resistant bladder cancer cells by cell cycle arrest, apoptosis and autophagy induction [39]. In present study, our results confirmed that autophagy inhibitor CQ could aggravate low dose of CDDP -inhibited cell viability, and silencing INE1 can aggravate low dose of CDDP-induced apoptosis of prostate cancer cells.
Precursor messenger RNA (pre-mRNA) splicing is an essential for gene expression from DNA to protein in all eukaryotes. Structural studies have proved that pre-mRNA is spliced by the spliceosome, which catalytic metals in the active site are coordinated by U6 small nuclear RNA and the catalytic triplex as a protein-directed metalloribozyme [40]. Aberrant mRNA splicing is prevalent in many cancers [41]. Dysregulation of spliceosome gene expression were investigated in advanced prostate cancer, such as SF1, HNRNPU, which are targeted by an RNA-binding transcriptional and splicing factor, splicing factor proline and glutamine-rich (PSF/SFPQ) [42]. LncRNA CTBP1-AS interacts with PSF and represses cell cycle regulators, and promotes castration-resistant prostate cancer growth [43]. It suggests that lncRNA-mediated association of RNA-binding proteins or transcription factors deregulates pre-mRNA splicing of spliceosome in prostate cancer progression, such as promoting cell proliferation, invasion, and drug resistance. In previous study, we demonstrated that lncRNA FAM66C activates epidermal growth factor receptor (EGFR)-extracellular signal-regulated kinase (ERK) signaling to promote cell proliferation by inhibiting proteasome pathway in prostate cancer [44]. Emerging research has also indicated that lncRNA acts as tumor oncogenes in human tumorigenesis by promoting anti-apoptosis of cancer cells [45, 46]. Our results suggest silencing INE1 increases apoptosis but does not affect cell proliferation in prostate cancer cell line DU145 and PC-3. SRSF2 is a splicing factor that binds to caspase-9 RNA [46], regulating the expression of its alternative spliced anti-apoptotic form [22, 23]. In particular, it has been reported that basal and cisplatin-induced caspase-9 activation and caspase-3 protein level is regulated by the splicing factor SRSF2 [47]. In the present study, silencing INE1 suppress the expression of SRSF2, which causes increasing the protein levels of caspase-9 and caspase-3, as well as cisplatin-induced DNA damage. Taken together, our study indicates that lncRNA INE1 plays an anti-apoptotic role by targeting the expression of the splicing factor SRSF2 in prostate cancer cell line DU145 and PC-3.
None.
Ethical policy
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study. Approval from institutional ethical committee was taken.
Availability of data and materials
The data sets supporting the conclusions of this study are included in this article and its additional images. Raw data are available on the main electronic data storage system of Affiliated Hospital of Jiangsu University, and access can be provided upon request to the authors.
Author contributions
HC and KL performed the research; HC, JG, YX and JM designed the research study and contributed essential reagents or tools; HC, KL, JG, WX, YX and JM analyzed and interpreted the data; HC, JG, WX, YX and JM wrote the manuscript. All authors reviewed the manuscript.
Competing interests
The authors declare no conflict of interest.
Funding
This research was funded by the Foundation of Health and Family Planning Commission of Wuxi City (Q201807), the Foundation of Clinical Medical Science and Technology Development project of Jiangsu University (JLY20180005), the Foundation of Science and Technology Development Medical and Health Guidance Plan Project of Wuxi City (2020061), the Innovation Foundation of Science and Technology Bureau of Yixing City (2020SF08), the Municipal Health Commission Suitable Technology promotion project of Wuxi City (T202126), and the Municipal Health Commission Suitable Technology promotion project of Jiangsu province (LSD2022008).
- Siegel RL, Miller KD, Jemal A: Cancer statistics, 2020. CA Cancer J Clin 2020, 70(1): 7-30.
- Etzioni R, Tsodikov A, Mariotto A, Szabo A, Falcon S, Wegelin J, DiTommaso D, Karnofski K, Gulati R, Penson DF, Feuer E: Quantifying the role of PSA screening in the US prostate cancer mortality decline. Cancer Causes Control 2008, 19(2): 175-181.
- Tsodikov A, Gulati R, Heijnsdijk EAM, Pinsky PF, Moss SM, Qiu S, de Carvalho TM, Hugosson J, Berg CD, Auvinen A, Andriole GL, Roobol MJ, Crawford ED, Nelen V, Kwiatkowski M, Zappa M, Lujan M, Villers A, Feuer EJ, de Koning HJ, Mariotto AB, Etzioni R: Reconciling the Effects of Screening on Prostate Cancer Mortality in the ERSPC and PLCO Trials. Ann Intern Med 2017, 167(7): 449-455.
- Negoita S, Feuer EJ, Mariotto A, Cronin KA, Petkov VI, Hussey SK, Benard V, Henley SJ, Anderson RN, Fedewa S, Sherman RL, Kohler BA, Dearmon BJ, Lake AJ, Ma J, Richardson LC, Jemal A, Penberthy L: Annual Report to the Nation on the Status of Cancer, part II: Recent changes in prostate cancer trends and disease characteristics. Cancer 2018, 124(13): 2801-2814.
- Giordano TJ: The cancer genome atlas research network: a sight to behold. Endocr Pathol 2014, 25(4): 362-365.
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM: The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015, 47(3): 199-208.
- Ponting CP, Oliver PL, Reik W: Evolution and functions of long noncoding RNAs. Cell 2009, 136(4): 629-641.
- Park JY, Lee JE, Park JB, Yoo H, Lee SH, Kim JH: Roles of Long Non-Coding RNAs on Tumorigenesis and Glioma Development. Brain Tumor Res Treat 2014, 2(1): 1-6.
- Shi X, Sun M, Liu H, Yao Y, Song Y: Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett 2013, 339(2): 159-166.
- Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA: Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol 2014, 65(6): 1140-1151.
- Qiu MT, Hu JW, Yin R, Xu L: Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol 2013, 34(2): 613-620.
- Ramnarine VR, Kobelev M, Gibb EA, Nouri M, Lin D, Wang Y, Buttyan R, Davicioni E, Zoubeidi A, Collins CC: The evolution of long noncoding RNA acceptance in prostate cancer initiation, progression, and its clinical utility in disease management. Eur Urol 2019, 76(5): 546-559.
- Goto T, Monk M: Regulation of X-chromosome inactivation in development in mice and humans. Microbiol Mol Biol Rev 1998, 62(2): 362-378.
- Li H, Yue H, Hao Y, Li H, Wang S, Yu L, Zhang D, Cao Y, Zhao J: Expression profile of long noncoding RNAs in human cerebral aneurysms: a microarray analysis. J Neurosurg 2017, 127(5): 1055-1062.
- Thiselton DL, McDowall J, Brandau O, Ramser J, d'Esposito F, Bhattacharya SS, Ross MT, Hardcastle AJ, Meindl A: An integrated, functionally annotated gene map of the DXS8026-ELK1 interval on human Xp11.3-Xp11.23: potential hotspot for neurogenetic disorders. Genomics 2002, 79(4): 560-572.
- Li J, Du H, Chen W, Qiu M, He P, Ma Z: Identification of potential autophagy-associated lncRNA in prostate cancer. Aging (Albany NY) 2021, 13(9): 13153-13165.
- Zheng X, Peng Q, Wang L, Zhang X, Huang L, Wang J, Qin Z: Serine/arginine-rich splicing factors: the bridge linking alternative splicing and cancer. Int J Biol Sci 2020, 16(13): 2442-2453.
- Chen Z, Hu H: Identification of prognosis biomarkers of prostatic cancer in a cohort of 498 patients from TCGA. Curr Probl Cancer 2019, 43(6): 100503.
- Wang H, Fang L, Jiang J, Kuang Y, Wang B, Shang X, Han P, Li Y, Liu M, Zhang Z, Li P: The cisplatin-induced lncRNA PANDAR dictates the chemoresistance of ovarian cancer via regulating SFRS2-mediated p53 phosphorylation. Cell Death Dis 2018, 9(11): 1103.
- Xu L, Wang Z, Yin C, Pan F, Shi T, Tian Y: Long noncoding RNA LINC02580 suppresses the invasion-metastasis cascade in hepatocellular carcinoma by targeting SRSF1. Biochem Biophys Res Commun 2020, 533(4): 685-691.
- Zhou D, Zhu X, Wu X, Zheng J, Tou L, Zhou Y: The effect of splicing MST1R in gastric cancer was enhanced by lncRNA FENDRR. Exp Ther Med 2021, 22(2): 798.
- Kedzierska H, Piekielko-Witkowska A: Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett 2017, 396: 53-65.
- Pellarin I, Dall'Acqua A, Gambelli A, Pellizzari I, D'Andrea S, Sonego M, Lorenzon I, Schiappacassi M, Belletti B, Baldassarre G: Splicing factor proline- and glutamine-rich (SFPQ) protein regulates platinum response in ovarian cancer-modulating SRSF2 activity. Oncogene 2020, 39(22): 4390-4403.
- Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, et al: The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet 2013, 45(11): 1392-1398.
- Lang C, Yin C, Lin K, Li Y, Yang Q, Wu Z, Du H, Ren D, Dai Y, Peng X: m(6) A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clin Transl Med 2021, 11: e426.
- Wen S, Wei Y, Zen C, Xiong W, Niu Y, Zhao Y: Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol Cancer 2020, 19(1): 171.
- Zhang B, Zhang M, Shen C, Liu G, Zhang F, Hou J, Yao W: LncRNA PCBP1-AS1-mediated AR/AR-V7 deubiquitination enhances prostate cancer enzalutamide resistance. Cell Death Dis 2021, 12(10): 856.
- Zhang Y, Guo S, Wang S, Li X, Hou D, Li H, Wang L, Xu Y, Ma B, Wang H, Jiang X: LncRNA OIP5-AS1 inhibits ferroptosis in prostate cancer with long-term cadmium exposure through miR-128-3p/SLC7A11 signaling. Ecotoxicol Environ Saf 2021, 220: 112376.
- Cai X, Dai Y, Gao P, Ren G, Cheng D, Wang B, Wang Y, Yu J, Du Y, Wang X, Xue B: LncRNA CCAT1 promotes prostate cancer cells proliferation, migration, and invasion through regulation of miR-490-3p/FRAT1 axis. Aging (Albany NY) 2021, 13(14): 18527-18544.
- Zhang N, Li Z, Bai F, Zhang S: PAX5-induced upregulation of IDH1-AS1 promotes tumor growth in prostate cancer by regulating ATG5-mediated autophagy. Cell Death Dis 2019, 10(10): 734.
- Fan L, Li H, Wang W: Long non-coding RNA PRRT3-AS1 silencing inhibits prostate cancer cell proliferation and promotes apoptosis and autophagy. Exp Physiol 2020, 105(5): 793-808.
- Cai Z, Wu Y, Ju G, Wang G, Liu B: Role of BCAR4 in prostate cancer cell autophagy. Transl Androl Urol 2021, 10(11): 4253-4261.
- Levy JMM, Towers CG, Thorburn A: Targeting autophagy in cancer. Nat Rev Cancer 2017, 17(9): 528-542.
- Jiang N, Meng C, Han X, Guo J, Li H, Yu Z: Low-dose cisplatin causes growth inhibition and loss of autophagy of rat astrocytes in vitro. Neurosci Lett 2018, 682: 112-117.
- Liu J, Chen Z, Guo J, Wang L, Liu X: Ambra1 induces autophagy and desensitizes human prostate cancer cells to cisplatin. Biosci Rep 2019, 39(8): BSR20170770.
- Zhu Q, Li H, Liu Y, Jiang L: Knockdown of CFTR enhances sensitivity of prostate cancer cells to cisplatin via inhibition of autophagy. Neoplasma 2017, 64(5): 709-717.
- Pennati M, Lopergolo A, Profumo V, De Cesare M, Sbarra S, Valdagni R, Zaffaroni N, Gandellini P, Folini M: miR-205 impairs the autophagic flux and enhances cisplatin cytotoxicity in castration-resistant prostate cancer cells. Biochem Pharmacol 2014, 87(4): 579-597.
- Peng L, Sang H, Wei S, Li Y, Jin D, Zhu X, Li X, Dang Y, Zhang G: circCUL2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142-3p/ROCK2. Mol Cancer 2020, 19(1): 156.
- Zhao F, Vakhrusheva O, Markowitsch SD, Slade KS, Tsaur I, Cinatl J Jr, Michaelis M, Efferth T, Haferkamp A, Juengel E: Artesunate Impairs Growth in Cisplatin-Resistant Bladder Cancer Cells by Cell Cycle Arrest, Apoptosis and Autophagy Induction. Cells 2020, 9(12): 2643.
- Shi Y: Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat Rev Mol Cell Biol 2017, 18(11): 655-670.
- Dvinge H, Kim E, Abdel-Wahab O, Bradley RK: RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer 2016, 16(7): 413-430.
- Takayama KI, Suzuki T, Fujimura T, Yamada Y, Takahashi S, Homma Y, Suzuki Y, Inoue S: Dysregulation of spliceosome gene expression in advanced prostate cancer by RNA-binding protein PSF. Proc Natl Acad Sci U S A 2017, 114(39): 10461-10466.
- Takayama K, Horie-Inoue K, Katayama S, Suzuki T, Tsutsumi S, Ikeda K, Urano T, Fujimura T, Takagi K, Takahashi S, et al: Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J 2013, 32(12): 1665-1680.
- Xie Y, Gu J, Qin Z, Ren Z, Wang Y, Shi H, Chen B: Long non-coding RNA FAM66C is associated with clinical progression and promotes cell proliferation by inhibiting proteasome pathway in prostate cancer. Cell Biochem Funct 2020, 38(8): 1006-1016.
- Dong HX, Wang R, Jin XY, Zeng J, Pan J: LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via inhibiting hsa-mir-22-3p. J Cell Physiol 2018, 233(5): 4126-4136.
- Wang J, Su Z, Lu S, Fu W, Liu Z, Jiang X, Tai S: LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta 2018, 485: 229-233.
- Merdzhanova G, Edmond V, De Seranno S, Van den Broeck A, Corcos L, Brambilla C, Brambilla E, Gazzeri S, Eymin B: E2F1 controls alternative splicing pattern of genes involved in apoptosis through upregulation of the splicing factor SC35. Cell Death Differ 2008, 15(12): 1815-1823.
Annals of urologic oncology
p-ISSN: 2617-7765, e-ISSN: 2617-7773
 Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript