Review Article | Open Access
Long Non-coding RNAs in Castration-resistant and Neuroendocrine Prostate Cancer: Potential Role and Therapeutic Impact
Kumari S. Prajapati1, Atul K. Singh1, Mohd Shuaib1, Prem P. Kushwaha1, Shashank Kumar1, Sanjay Gupta2, 3, 4, 5, 6
1Molecular Signaling & Drug Discovery Laboratory, Department of Biochemistry, Central University of Punjab, Bathinda 151401, India.
2Department of Urology, Case Western Reserve University, Cleveland, Ohio 44106, USA.
3The Urology Institute, University Hospitals Cleveland Medical Center, Cleveland, OH 44106, USA.
4Department of Urology, Louis Stokes Cleveland Veterans Affairs Medical Center, Cleveland, OH 44106, USA.
5Department of Nutrition, Case Western Reserve University, Cleveland, OH 44106, USA.
6Division of General Medical Sciences, Case Comprehensive Cancer Center, Cleveland, OH 44106, USA.
Correspondence: Sanjay Gupta (Department of Urology, The James and Eilleen Dicke Research Laboratory, Case Western Reserve University, 10900 Euclid Avenue, Cleveland, Ohio 44106 USA; Email: sanjay.gupta@case.edu).
Annals of Urologic Oncology 2020, 3(2): 71-81. https://doi.org/10.32948/auo.2020.12.03
Received: 28 Nov 2020 | Accepted: 29 Nov 2020 | Published online: 31 Dec 2020
Prostate cancer is the most commonly diagnosed malignancy and leading cause of cancer-related deaths in men worldwide. The disease is heterogeneous in nature exhibiting various clinical subtypes and genetic/transcriptomic features. Long non-coding RNAs (lncRNAs) are transcripts of more than 200 nucleotides that does not encode any protein and play important role in several biological processes as well as pathologic states. Deregulation of lncRNAs has been associated with human diseases. In prostate cancer, numerous key lncRNAs have been identified as novel players that contribute to the pathophysiology of the disease primarily regulated by androgen and its cognate receptor. The present review attempts to summarize the potential role of lncRNA and their mechanisms of action in prostate cancer with particular focus on lncRNAs regulated by androgen receptor expressed in castration-resistant and neuroendocrine differentiated subtypes. We also emphasize the potential of these lncRNAs for their development as therapeutic targets in prostate cancer.
Key words long non-coding RNA, castration-resistant prostate cancer, neuroendocrine prostate cancer, androgen signaling
Prostate cancer is the second-leading and most common cause of cancer-related death in the male population worldwide [1]. Prostate cancer progression occurs from androgen-dependent to androgen-independent stage and acquires metastatic capabilities at the advanced-stage malignancy. Treatment of prostate cancer differs at various stages of progression. Hormone-sensitive prostate tumors that are organ-confined are treated by surgery or radiotherapy whereas non-organ-confined tumors are subjected to hormonal therapies targeting the androgen receptor (AR) axis. Majority of patients experience tumor remission on androgen deprivation therapy (ADT), however, ~30% of these tumors reemerge and are clinically defined as castration-resistant prostate cancer (CRPC). A study reported that within 5 years, 10% to 20% of prostate cancer patients develop CRPC on ADT treatment [2]. Clinical use of second-generation drugs which target AR axis including enzalutamide and abiraterone acetate results in the development of CRPC phenotype. The ineffectiveness of ADT as a result of chemoresistance results in the activation of AR and its downstream pathways in CRPC tumors. Other key factors responsible for the CRPC transition include intratumoral androgen production, AR ligand-binding domain activation, AR amplification, AR splice variants formation, overexpression of feedback pathways [3]. In addition to the change in CRPC features, the dynamics of cellular plasticity alters during cancer progression, where tumor cells transition to AR-independence attaining neuroendocrine characteristics [4]. Neuroendocrine prostate cancer (NEPC) is the most lethal and aggressive form possessing an unusual phenotype. The median survival of NEPC patients is < 7 months and platinum-based chemotherapy remains the solitary treatment option [5]. It is still unclear whether NEPC tumors develop either de novo or as an adaptive response to ADT possessing aggressive pathological appearance with poor clinical outcome and overall low survival [6, 7].
Numerous reports suggest that AR and its downstream signaling are fundamental for the development and progression of both localized and advanced-stage prostate cancer. AR is a ligand-dependent transcriptional factor and a member of the steroid hormone nuclear receptor family. AR shares structural and functional homology with progesterone, estrogen, mineralocorticoid, and glucocorticoid receptors [8, 9]. The human AR is located on chromosome X (Xq11-12) and consists of eight exons and seven introns [10]. AR is a 90-kb gene encoding 110-kDa molecular weight protein containing 919 amino acids. AR is organized into four distinct functional domains including the Ligand-binding domain (LBD), DNA-binding domain (DBD), Hinge domain, and amino-terminal domain (NTD) [11]. Upon binding of ligands, androgens such as dihydrotestosterone and testosterone change the AR conformation to an active state. AR is normally present in an inactive state bound with heat shock proteins (HSP70 and HSP90) and with chaperone proteins in the cytosol. The binding of androgens triggers the release of HSP from the AR necessary for dimerization, translocation into the nucleus and binding to specific androgen response elements (AREs), regulating target gene transcription and expression. AR transcript expression is also mediated with the support of other coactivators and co-suppressor proteins [12]. Identification of AR downstream molecules and novel regulators for AR activation are important in understanding the mechanism(s) of CRPC and NEPC and to improve the treatment of advanced prostate cancer.
The recent development of high-throughput technology has led to the identification of noncoding transcripts, such as long noncoding RNAs (lncRNAs). Aberrant expression of lncRNA has been noted in a variety of human diseases including cancer, contributing to the pathogenesis or maintaining diseased conditions. The present review summarizes the current knowledge of lncRNAs expression patterns in advance-stage prostate cancer including its role during malignant progression, tumor suppression, and towards the development of chemo-resistance.
Recent advancement and improvements in the therapeutic strategy to suppress metastatic CRPC with second-generation AR-targeting agents with enzalutamide and abiraterone acetate increases patient survival by approximately 4.8 months [13]. Studies suggest that alteration of AR-signaling pathways and AR-signaling bypass mechanisms leads to the development of CRPC whereas the mechanism(s) of AR-independent clonal evolution promotes the emergence of NEPC tumors [14]. Some of the AR bypass mechanisms include glucocorticoid receptor, PI3K-AKT-mTOR pathway, and stress signaling [15]. Increasing evidence demonstrates a positive association between CRPC and NEPC tumors [16-18]. Clinical data suggested that following the first and second-line therapy targeting AR-axis, NEPC is found to be present in 30-40% of patients with metastatic CRPC [19]. Recent studies corroborated that continuous exposure and selective pressure of ADT induces neuroendocrine trans-differentiation, resulting in the emergence of NEPC tumors [14].
CRPC and NEPC share similar genomics but different transcriptomics [20]. Mu et al. (2017) reported that selection pressure mediated by AR-axis inhibitors stimulates NEPC formation in prostate cancer cells along with the loss of TP53 and RB1 expression [21]. Inactivation of RB1and TP53 induces the expression of the SOX2 transcription factor to entrap on lineage plasticity in prostate cancer cells and reprogramming of fibroblasts to induce-pluripotent stem cells [21]. Another important protein overexpressed under the influence of TP53 and RB1 inactivation is Enhancer of zeste homolog 2 (EZH2) [22]. EZH2, the master regulator of neuroendocrine lineage plasticity was observed to be upregulated in NEPC patients where EZH2 inhibitors are able to restore AR sensitivity [22]. Another study investigated that overexpression of N-Myc with upregulated AKT drives the formation of NEPC in mice models [23, 24]. Dardenne et al. (2016) reported that activated Aurora A kinase (AURKA) increases cell proliferation in NEPC in the absence of AR and amplification of MYCN collaborate with EZH2 to reduces AR transactivation, and promote neuroendocrine differentiation [23]. The study also reported that 40% of NEPC cells have upregulated AURKA and amplified MYCN [25]. One of the transcription factors, repressor element 1 (RE-1) silencing transcription factor (REST) particularly known as a negative master regulator of neuronal differentiation plays a critical role in cell differentiation fate [26]. REST is highly expressed in prostate adenocarcinoma and found to be downregulated in NEPC patients [27, 28]. Downregulation of REST leads to the repression of the neuronal-differentiation-related genes which ultimately leads to NEPC development. AR-axis inhibitors regulate REST both at the mRNA and protein level. REST is also regulated by SRRM4, β-TRCP and HAUSP to maintain it in an inactive form. Downregulation of REST through AR-axis inhibition via the upregulation of SRRM4 and β-TRCP induces neuroendocrine-specific differentiation among prostate cancer cells [14]. Studies also demonstrate the involvement of placental genes such as PEG10, FOXA1, and BRN2 to promote and activate neuroendocrine differentiation in prostate cancer cells [29]. HIF1A is reported to regulate the neuroendocrine characteristics in prostate cancer cells. HIF1A in cooperation with the transcription factor FOXA2 induces several HIF1A target genes essential for promoting hypoxia-induced metastasis and neuroendocrine phenotype in prostate cancer [30-32].
Recent studies reported the role of ONECUT2 overexpression in inhibiting AR signaling and promoting neuroendocrine phenotype under hypoxic conditions [31, 33]. ONECUT2 plays an important role in contributing to angiogenic phenotype in NEPC. It is also demonstrated that NEPC tumors exhibit various developmental programs related to epithelial-to-mesenchymal (EMT) induction and stem-like characteristics. Dicken et al. (2018) demonstrated that the signaling interaction between AR axis and TGF-β mechanistic network influences the EMT phenotypic transformations creating the critical environment for epithelial-derived tumors to become more invasive to adopt neuroendocrine features [34]. The involvement of causal pathways facilitating CRPC and NEPC initiation and development is shown in Figure 1.
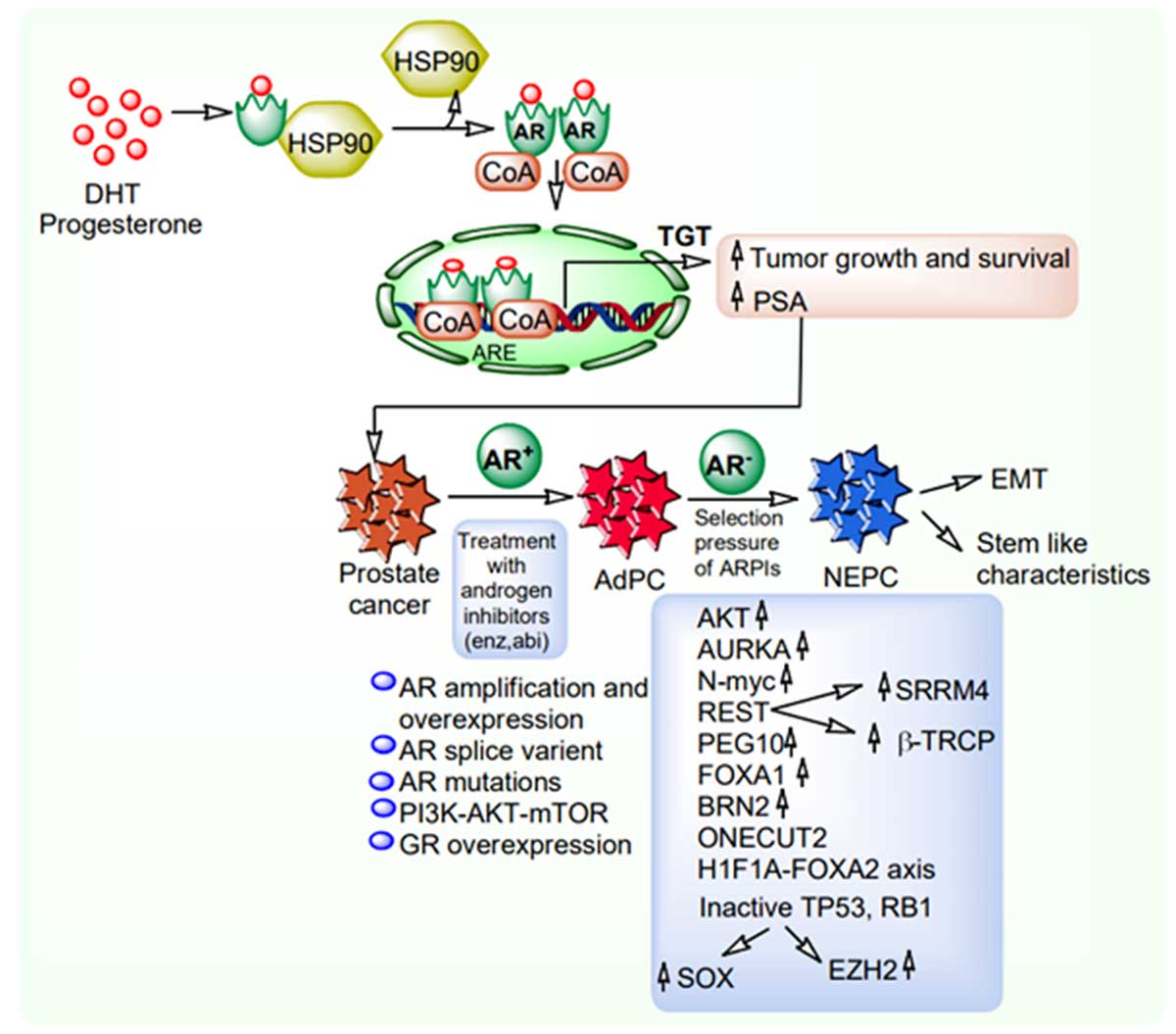 Figure 1. Molecular signaling of prostate cancer, castration resistant prostate cancer (CRPC) and neuroendocrine prostate cancer (NEPC)
progression. DHT: Dihydrotestosterone; HSP90: Heat shock protein 90; AR: Androgen receptor; CoA: Coenzyme A; TGT: Target gene
transcription; ARE: Androgen response element; PSA: Prostate serum antigen; Enz: Enzalutamide; Abi: Abiraterone acetate; AdPC:
Adenocarcinoma prostate cancer; ARPI: Androgen receptor pathway inhibitor; NEPC: Neuroendocrine prostate cancer; EMT: Epithelial
mesenchymal transition; AKT: Protein kinase B; AURKA: Aurora kinase A; REST: RE1-Silencing transcription factor; PEG10: Paternally
expressed 10; FOXA1: Forkhead Box1; HIF1A: Hypoxia inducing factor 1A; TP53: Tumor suppressor protein 53; RB1: Retinoblastoma1; SOX:
SRY-related HMG-box; EZH2: Enhancer of zeste homolog 2; PI3K: Phosphoinositide 3-kinase; mTOR: Mammalian target of rapamycin;
SRRM4: Serine/arginine repetitive matrix 4; β-TRCP: Beta transducing repeats-containing proteins.
Figure 1. Molecular signaling of prostate cancer, castration resistant prostate cancer (CRPC) and neuroendocrine prostate cancer (NEPC)
progression. DHT: Dihydrotestosterone; HSP90: Heat shock protein 90; AR: Androgen receptor; CoA: Coenzyme A; TGT: Target gene
transcription; ARE: Androgen response element; PSA: Prostate serum antigen; Enz: Enzalutamide; Abi: Abiraterone acetate; AdPC:
Adenocarcinoma prostate cancer; ARPI: Androgen receptor pathway inhibitor; NEPC: Neuroendocrine prostate cancer; EMT: Epithelial
mesenchymal transition; AKT: Protein kinase B; AURKA: Aurora kinase A; REST: RE1-Silencing transcription factor; PEG10: Paternally
expressed 10; FOXA1: Forkhead Box1; HIF1A: Hypoxia inducing factor 1A; TP53: Tumor suppressor protein 53; RB1: Retinoblastoma1; SOX:
SRY-related HMG-box; EZH2: Enhancer of zeste homolog 2; PI3K: Phosphoinositide 3-kinase; mTOR: Mammalian target of rapamycin;
SRRM4: Serine/arginine repetitive matrix 4; β-TRCP: Beta transducing repeats-containing proteins.
Various lncRNAs are known to play major roles in CRPC progression. These lncRNAs regulate key cellular processes in CRPC cells such as cell proliferation, invasion, metastasis, angiogenesis, and resistance to apoptosis [41]. Genome-wide association studies have revealed a high proportion of genetic alterations in prostate cancer-associated with lncRNAs coding region of the genome, suggesting the important role of lncRNAs in cancer progression [42, 43]. The intergenic region 8q24 is a gene desert located near the MYC proto-oncogene is reported to be involved in carcinogenesis [44]. In prostate cancer, 8q24 is a major susceptibility region that transcribes various single nucleotide polymorphism containing RNAs that are involved in the promotion of hormone-dependent prostate cancer to CRPC [45]. lncRNAs such as prostate-specific transcript 1 (PCGEM1), prostate cancer-associated non-coding RNA 1 (PRNCR1), and steroid receptor RNA activator (SRA) are reported to enhance AR-mediated transcription of target genes via a chromatin loop formation. These lncRNAs bind to the enhancer region of AR target genes and recruit DOT1L methyltransferase which promotes AR methylation on K349. Methylation of AR on K349 results in the recruitment of pygo 2 which further enhances loop formation between enhancer and promoter resulting in elevated transcription of the target genes. Furthermore, lncRNAs mediated loop formation promotes prostate cancer progression via both ligand-dependent and ligand-independent manner [46]. Apart from prostate cancer progression by PCGEM1 via loop formation mechanism, it also promotes cancer progression via upregulation of metabolism [47]. PCGEM1 promotes CRPC metabolically through the upregulation of aerobic glycolysis, pentose phosphate pathway, lipid and glutamine metabolism. Regulation of metabolism by PCGEM1 is independent of the presence of AR and androgen and it is mediated by MYC activation [47] (Figure 2). Another lncRNAs reported to associate with tumor metabolism is lincRNA-p21. Under the hypoxic condition, hypoxia-inducible factor α (HIF-α) induces the expression of lincRNAp21. Isin et al. (2015) reported the elevated expression of lincRNA-p21 in prostate cancer suggests lincRNA-p21 could be an important player in the regulation of CRPC metabolism [48].
The lncRNA HOX transcript antisense RNA (HOTAIR) is reported to facilitate AR-mediated transcriptional response and promote CRPC [49]. Another mechanism by which HOTAIR regulates CRPC progression is by reciprocal regulation of EZH2 and DNMT1 which results in the polyphylline-1 inhibited progression of CRPC [50]. HOTAIR protects AR from E3 ubiquitin ligase MDM2-mediated protein degradation through direct interaction with AR. Therefore, overexpression of HOTAIR contributes to upregulates AR target gene expression in an androgen-independent mode and thus promotes CRPC progression [49] (Figure 3). The lncRNA suppressor of cytokine signaling 2- antisense transcript 1 (SOCS2-AS1) also facilitates AR-mediated transcriptional response. SOCS2-AS1 associated with AR protein promotes transcriptional repression of tumor suppressor gene TNFSF10 [51] (Figure 4).
Prostate-specific lncRNA prostate cancer-associated transcript 1 (PCAT1) is involved in the regulation of stress response and DNA repair processes. Uncontrolled cell division of cancer cells results in the accumulation of many types of aberrations in the DNA of cancer cells which leads to genomic instability. PCAT1 was reported to be upregulated in CRPC and it activates AKT and NF-?B signaling pathways by regulating PHLPP/FKBP51/IKKα complex [52]. Prensner et al. (2014) reported that PCAT1 downregulates tumor suppressor gene BRCA1 results in aberrant homologous recombination and DNA repair [53]. PCAT1 is associated with PRC2 subunit, suppressor of zeste 12 homolog (SUZ12), and functions as a transcriptional repressor [54].
Other lncRNAs involve in the progression of CRPC are Nuclear enriched abundant transcript 1 (NEAT1) and Prostate cancer-associated transcript 5 (PCAT5). NEAT1 regulates the expression level of specific prostate cancer-associated genes along with the modulation of expression of TMPRSS2-ERG fusion transcripts [55]. PCAT5 is overexpressed in ERG positive CRPCs. PCAT5 promotes proliferation, invasion and resistance to apoptosis in CRPC cells [56]. SChLAP1 is another lncRNA overexpressed in CRPC. SWI/SNF Complex antagonist associated with prostate cancer 1 (SChLAP1) promotes invasion and migration of CRPC cells by regulating the SWItch/Sucrose Non-Fermentable-complex and inhibiting its gene expression regulatory function [57].
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) lncRNA is reported to be involved with pathological complications of organs that have a strong association with sex hormones. MALAT1 is one of the 10 most upregulated transcripts in CRPC and its upregulation is associated with the CRPC progression [58, 59]. Wang et al. (2015) reported that MALAT1 regulates the activity of EZH2 in CRPC via facilitating promoter activity and H3K27me3 activity of EZH2 [60]. MALAT1 cooperates with EZH2 playing a crucial role in EZH2-mediated migration and invasion in CRPC cell lines [60]. Studies reported the high expression of MALAT1 in CRPC whose knockdown reduces cell growth and invasion which are also repressed by androgen treatment.
Growth arrest-specific 5 (GAS5) lncRNA is reported to be downregulated in mCRPC and the result of which shows reduced chemotherapy-induced cellular apoptosis [61]. It is apparent that low-level GAS5 makes mTOR inhibitors insensitive to CRPC cells [62].
It has been demonstrated that lncRNAs function as a decoy or molecular sponge for the miRNAs targeting protein-coding mRNA. Zhu et al. (2014) demonstrated the significant downregulation of H19 and H19-derived miR-675 in metastatic prostate cancer cells as compared to non-metastatic prostate cancer cells [63]. However, the upregulation of H19 increases miR-675 expression inhibiting prostate cancer cell migration. Additionally, miR-675 targets RB and CDC6, residing within the first exonic region of H19 [64, 65]. H19 and miR-675 are significantly downregulated that increases the expression of TGF-β1 thus promoting cell migration. Binding of miR-675 at the 3’ UTR of TGF-β1 mRNA inhibits its translation, thereby H19-miR-675 axis considered as a suppressor of metastatic prostate cancer [63]. Upregulated lncRNA MIR221HG which is transcribed in the promoter region of miR-221/222 is reported to promote cell growth in an androgen-independent manner. Upregulation of MIR221HG inhibits the expression of CRPC markers including TMPRSS2, KLK3, and FKBP5 produced on AR induction. Clinically, a high expression level of MIR222HG is reported with prostate cancer progression and development to CRPC [66].
PTENP1, a pseudogene of the tumor suppressor gene PTEN containing MREs transcript similar to PTEN targeting miRNAs including miR-21, miR-124, miR-17, miR-26a, and miR-19 families [67]. Furthermore, PTEN loss is greatly associated with the development of CRPC and with poor clinical outcomes in CRPC patients [68]. PTENP1 and PTEN are positively correlated to each other in prostate cancer progression. PTENP1 and PTEN compete with each other for binding to their common miRNAs. Overexpression of 3’ UTR PTENP1 diminished the proliferation rate of DU145 cells through the upregulation of PTEN both at the mRNA a protein levels [69]. Therefore, PTENP1 acts as a decoy through altering anti-PTEN miRNA away from PTEN and the result of which increases tumor suppressor activity of PTEN [67].
Clinical data suggest that NEPC arise de novo in prostate cancer patients with 0.5% to 2% frequency. Ramnarine et al. (2018) developed a first-in-field patient-derived xenograft (PDX) model of NEPC to study the clinical implications of the landscape of lncRNA in NEPC [70]. Longitudinal deep transcriptomic profiling of PDX of androgen deprivation enables to observe dynamic transcriptional changes during neuroendocrine differentiation. The group identified 122 lncRNAs that strongly distinguish the existence of NEPC different from adenocarcinoma tumor patients and the top identified lncRNAs are H19, LINC00617, FENDRR, LINC00514, and SSTR5-AS1 [70].
Transcription factor binding site (TFBS) analysis identified NEPC-associated TF motifs that are present within the NEtD lncRNA sequences which indicates the functional role of identified lncRNAs in NEPC pathogenesis [70]. FOXF1 adjacent non-coding developmentary regulatory RNA (FENDRR) is the most deregulated lncRNA in NEPC [71]. FENDRR is reported to interact with PRC2 which is a key player in prostate tumor progression. FENDRR is reported to reduce tumor invasion by targeting CSNK1E in PC-3 cells [72].
LINC00514 is a highly expressed lncRNA NEPC which is predicted to bind with TADA3 and lowers its activity [70]. TADA3 is an important regulator of cell proliferation through the stabilization and regulation of p53 activity [73]. The interaction between TADA3 and p53 could be one of the reasons for the loss of p53 in NEPC.
SSTR5 antisense RNA 1 (SSTR5-AS1) lncRNA was reported to be highly expressed in NEPC. SSTR5-AS1 is an antisense transcript of SSTR5 which belongs to the family of somatostatin receptor family [70]. Somatostatins are reported to regulate important cellular processes such as cell proliferation neurotransmission, and endocrine signaling [74]. Another lncRNA involved in the progression of NEPC is NEAR1. NEAR1 is reported to promote the proliferation and survival of NEPC cells [75].
Long intergenic non-coding protein-coding RNA 8 (H19) is an imprinted maternally expressed untranslated mRNA mainly located in the region of chromosome 11 (11p15.5) near the insulin-growth factor 2 (IGF2) gene [76]. In the case of metastatic prostate cancer, H19 plays a tumor-suppressive role by suppressing transforming growth factor-β1 (TGF-β1) effects [63]. Downregulation of H19 inhibited glucose metabolism, lactic acid production, cell growth, and proliferation in AR-negative prostate cancer cells [77].
The lncRNA-p21 was identified to activate p21 and a p53 corepressor to stimulate and promote the PRC2 target gene expression that affects cellular epigenetic regulation [78]. Luo et al. (2019) reported high expression of lncRNA-p21 in NEPC derived xenograft tissues [79]. Chang et al. (2018) proposed HOTAIR as a REST-regulated lncRNA that drives neuroendocrine differentiation in prostate cancer cells [80]. HOTAIR is identified as a novel REST-repressed and upregulated lncRNA in NEPC cells, whose knockdown suppresses neuroendocrine differentiation. HOTAIR is reported to interact with PRC2 complex and EZH2 allowing the repressor complex to bind to AR suppressing AR activity [81]. A study reported that SChLAP1 exerts its oncogenic effect by negatively regulating miR-198 thereby affecting the MAPK1 signaling pathway [82]. SChLAP1 is reported to be regulated in both CRPC and NEPC tumors. Thus, there is a possibility of involvement of SChLAP1 and miR-198 relation in prostate cancer progression to NEPC. Similarly, MALAT1 was also found to be negatively regulated by miR-1 expression in androgen receptor-negative prostate cancer cells. MALAT1 acts as a molecular sponge of miR-1 which increases the expression level of KRAS thereby affecting cell proliferation, apoptosis, and migration [83]. Ling et al. (2017) reported HOTAIR as a direct negative target of miR-193a [84]. MiR-193a performs tumor-suppressive functions, however, was found downregulated in metastatic prostate cancer through HOTAIR mediated EZH2 expression. Therefore, HOTAIR/EZH2/miR-193a feedback loop formation paying a vital role in NEPC progression [84].
LncRNA myocardial infarction associated transcript (MIAT) interacts with polycomb genes and is highly upregulated in NEPCs. Clinical database analysis by Crea et al. (2016) suggested higher expression of lncRNA MIAT is involved in prostate cancer progression and trans-differentiation from prostatic adenocarcinomas to NEPC [85]. LncRNA MIAT expression is confined to a small proportion of prostate cancer with recurrence in Rb mutations, high metastatic potential, and poor prognosis [85] (Table 1).
| Table 1. Represents various lncRNAs involved in CRPC and NEPC formation. | ||||
| lncRNAs | Cancer type | Target | Functions | References |
| PCGEM1 | CRPC | MYC | Increase aerobic glycolysis, PPP, lipid and glutamine metabolism, promotes CRPC | [47] |
| HOTAIR | CRPC, NEPC | EZH2, DNMT, REST | CRPC progression, Neuroendocrine differentiation | [50, 80] |
| SOCS2-AS1 | CRPC | TNFSF10 | Repression of tumor suppressor gene | [51] |
| lincRNA-p21 | CRPC | HIF-α | CRPC progression | [48] |
| PCAT1 | CRPC | AKT, NF-?B, PHLPP/FKBP51/IKKα complex, BRCA1 | Genomic instability, aberrant homologues recombination and DNA repair | [52] |
| NEAT1 | CRPC | TMPRSS2-ERG fusion transcripts | CRPC progression | [55] |
| PCAT5 | CRPC | ERG | Promotes proliferation, invasion and resistance to apoptosis in CRPC cells | [56] |
| SChLAP1 | CRPC | SWItch/Sucrose Non-Fermentable-complex | Promotes invasion and migration of CRPC | [57] |
| MALAT1 | CRPC | EZH2 | CRPC progression, migration and invasion in CRPC cells | [60] |
| GAS5 | mCRPC | mTOR | Reduced chemotherapy-induced cellular apoptosis | [61, 62] |
| FENDRR | NEPC | PRC2 | Tumor progression | [72] |
| LINC00514 | NEPC | TADA3, p53 | NEPC development | [73] |
| H19 | NEPC | TGF-β1 | Lactic acid production, cell growth, and proliferation in AR-negative prostate cancer cells | [63] |
| LncRNA-p21 | NEPC | PRC2 | Affect epigenetic regulation, NEPC formation | [78] |
| MIAT | NEPC | Polycomb genes | Trans-differentiation from prostatic adenocarcinomas to NEPC, high metastatic potential, and poor prognosis | [85] |
| CRPC, Castration resistant prostate cancer, NEPC, Neuroendocrine prostate cancer, PCGEM1, Prostate specific transcript 1; HOTAIR, HOX transcript antisense RNA; SOCS2-AS1, Suppressor of cytokine signaling 2- antisense transcript 1; PCAT1, Prostate cancer associated transcript 1; PCAT5, Prostate cancer associated transcript 5; MALAT1, Metastasis associated lung adenocarcinoma transcript 1; GAS5, Growth arrest-specific 5; FENDRR, FOXF1 adjacent non-coding developmentary regulatory RNA; MIAT, Myocardial infarction associated transcript; AKT, Protein kinase B; REST, RE1-Silencing transcription factor; HIF1A, Hypoxia inducing factor 1A; TP53, Tumor suppressor protein 53; EZH2, Enhancer of zeste homolog 2; mTOR, Mammalian target of rapamycin; PRC2, Polycomb repressive complex 2; TADA3, Transcriptional adaptor 3; DNMT, DNA methyl transferase; TGF-β1, Transforming growth factor-β1; TMPRSS2, Transmembrane protease serine 2; ERG, ETS-Related gene; NF-kb, Nuclear factor- kb; PHLPP, PH domain leucine-rich repeat protein phosphatase; FKBP5, FK506-Binding protein 51; IKKα, I?B Kinase α; BRCA1, Breast cancer type 1 susceptibility protein. | ||||
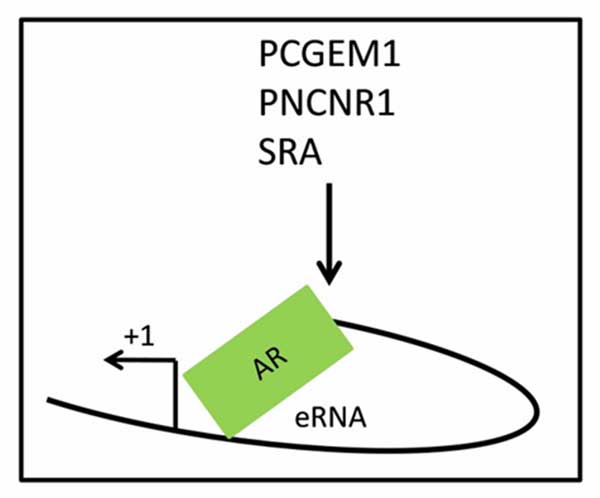 Figure 2. Regulation of AR expression by lncRNAs via promoter/
enhancer loop formation. Several lncRNAs such as PCGEM1,
PNCNR1, and SRA promote the promoter/enhancer loop formation
on AR regulated genes and facilitate the AR regulated gene
expression. PCGEM1: Prostate cancer gene expression marker 1;
PNCNR1: Prostate cancer noncoding RNA 1; SRA: Steroid receptor
RNA activator; AR: Androgen receptor protein; eRNA: Enhancer-
templated non-coding RNA.
Figure 2. Regulation of AR expression by lncRNAs via promoter/
enhancer loop formation. Several lncRNAs such as PCGEM1,
PNCNR1, and SRA promote the promoter/enhancer loop formation
on AR regulated genes and facilitate the AR regulated gene
expression. PCGEM1: Prostate cancer gene expression marker 1;
PNCNR1: Prostate cancer noncoding RNA 1; SRA: Steroid receptor
RNA activator; AR: Androgen receptor protein; eRNA: Enhancer-
templated non-coding RNA.
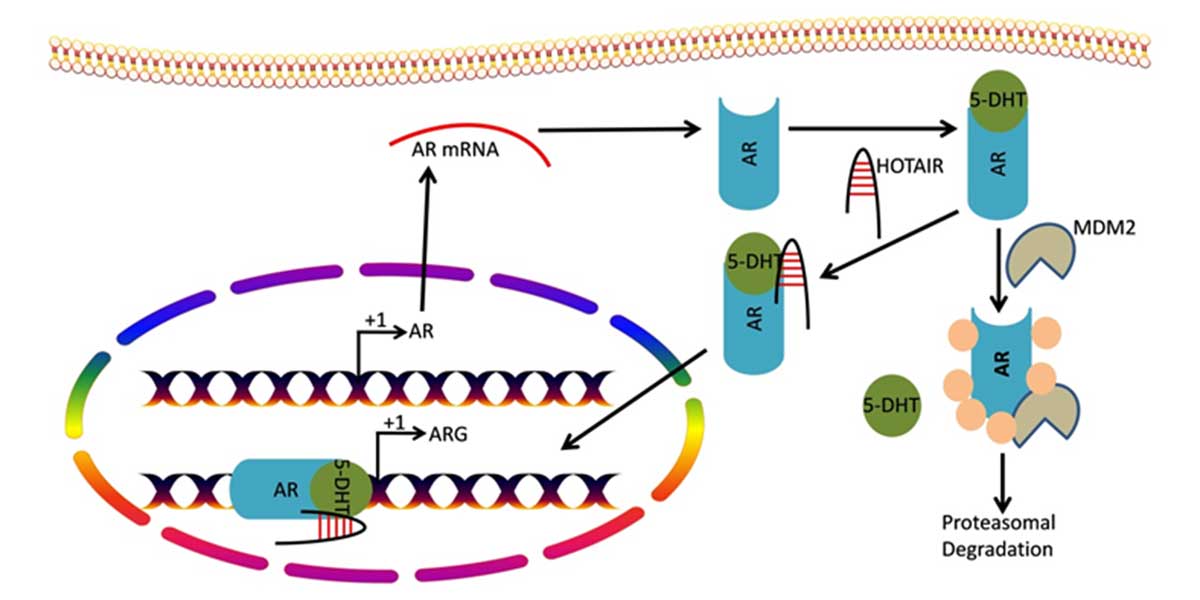 Figure 3. Inhibition of AR protein degradation by HOTAIR. E3 ubiquitin ligase MDM2 binds with AR protein and promotes its proteasomal
degradation. lncRNA HOTAIR binds with AR protein and prevents the binding of MDM2 and thereby promotes the AR mediated gene
expression. AR: Androgen receptor protein; 5-DHT: Dihydrotestosterone; ARG: AR responsive genes; HOTAIR: HOX transcript antisense
RNA; MDM2: Mouse double minute 2 homolog.
Figure 3. Inhibition of AR protein degradation by HOTAIR. E3 ubiquitin ligase MDM2 binds with AR protein and promotes its proteasomal
degradation. lncRNA HOTAIR binds with AR protein and prevents the binding of MDM2 and thereby promotes the AR mediated gene
expression. AR: Androgen receptor protein; 5-DHT: Dihydrotestosterone; ARG: AR responsive genes; HOTAIR: HOX transcript antisense
RNA; MDM2: Mouse double minute 2 homolog.
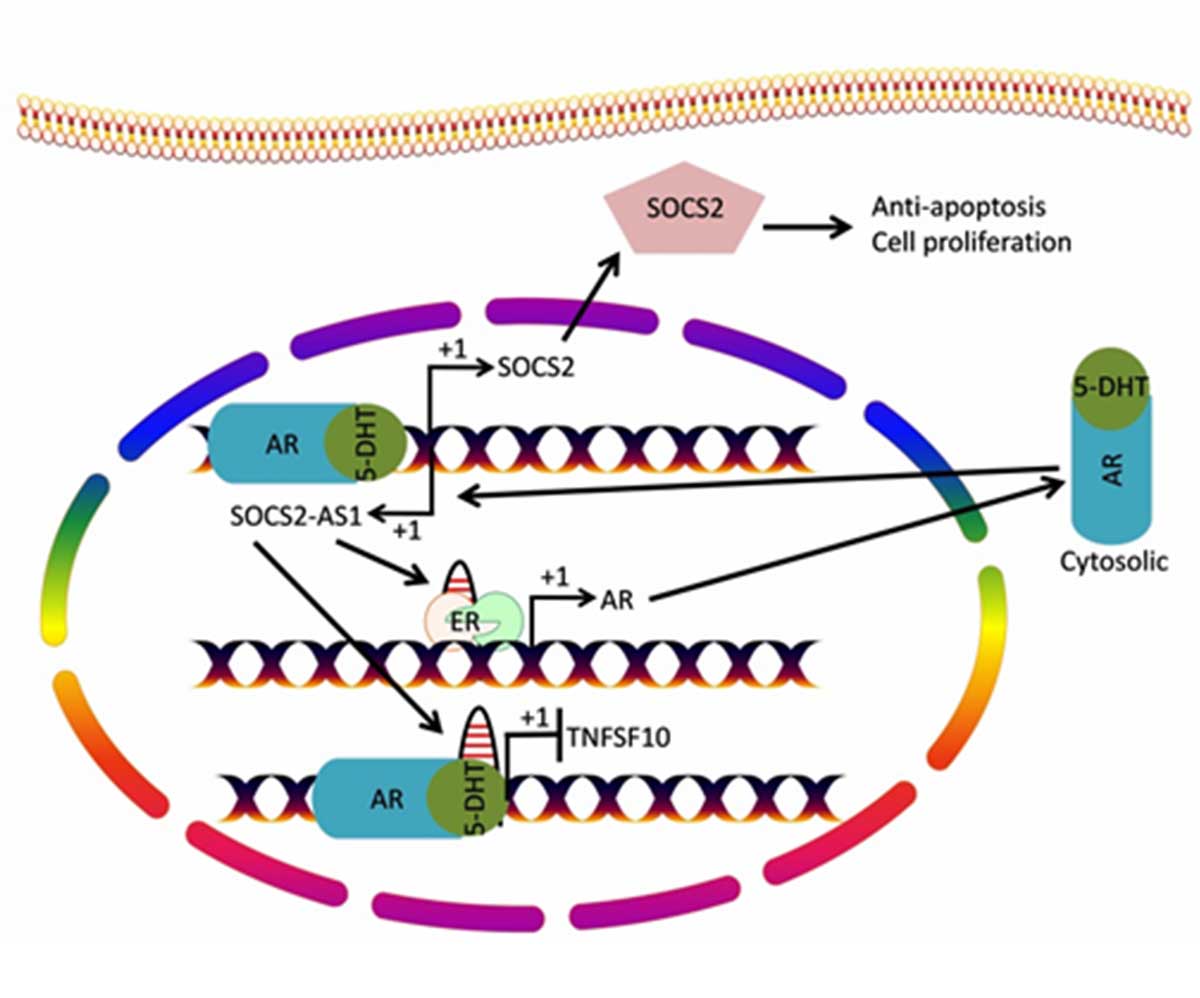 Figure 4. Interaction of SOCS2-AS1 lncRNA with AR. Cytosolic AR protein translocates into the nucleus and facilitates transcription of SOCS2
gene. Transcription of sense strand results in the production of SOCS2 protein which promotes carcinogenesis of prostate cancer via promoting
cell proliferation and anti-apoptosis. Transcription of antisense strand produces lncRNA SOCS2-AS1 which binds to AR protein and promotes
gene regulation such as transcriptional inhibition of tumor suppressor gene TNFSF10. Furthermore SOCS2-AS1 also promotes expression of
AR protein by directly binding to its promoter and recruiting epigenetic regulators on AR promoter. AR: Androgen receptor protein; 5-DHT:
Dihydrotestosterone; ER: Epigenetic regulators; SOCS2: Suppressor of Cytokine Signaling 2; SOCS2-AS1: Suppressor of cytokine signaling
2-antisense transcript 1; TNFS10: TNF Superfamily Member 10.
Figure 4. Interaction of SOCS2-AS1 lncRNA with AR. Cytosolic AR protein translocates into the nucleus and facilitates transcription of SOCS2
gene. Transcription of sense strand results in the production of SOCS2 protein which promotes carcinogenesis of prostate cancer via promoting
cell proliferation and anti-apoptosis. Transcription of antisense strand produces lncRNA SOCS2-AS1 which binds to AR protein and promotes
gene regulation such as transcriptional inhibition of tumor suppressor gene TNFSF10. Furthermore SOCS2-AS1 also promotes expression of
AR protein by directly binding to its promoter and recruiting epigenetic regulators on AR promoter. AR: Androgen receptor protein; 5-DHT:
Dihydrotestosterone; ER: Epigenetic regulators; SOCS2: Suppressor of Cytokine Signaling 2; SOCS2-AS1: Suppressor of cytokine signaling
2-antisense transcript 1; TNFS10: TNF Superfamily Member 10.
High levels of HOTAIR endorses CRPC progression. Polyphyllin I is a steroidal saponin that exhibited antitumor activity and inhibits the growth of CRPC cells by reducing the level of HOTAIR [88]. Paclitaxel, a natural anticancer drug, negatively regulates the expression of highly expressed lncRNA DANCR in prostate cancer tissues and cells, and its downregulation is associated with loss of proliferation in prostate cancer [89].
Analysis of prostate cancer cell lines and clinical specimens demonstrated upregulation of lncRNA-p21 in NEPC where treatment with enzalutamide was shown to increase lncRNA-p21 activity promoting neuroendocrine differentiation. This mechanistic study corroborated that enzalutamide treatment promotes transcription of lncRNA-p21 altering AR levels and it’s binding with AREs, thereby switching the function of EZH2 from histone-methyltransferase to non-histone methyltransferase. The switching mechanism of EZH2 methylates STAT3 to promote neuroendocrine differentiation [79]. Enzalutamide inhibitor GSK503 reported suppressing neuroendocrine differentiation [90]. Preclinical studies of EZH2 inhibitors such as GSK126 and DZNep reported to target AR/lncRNA-p21/EZH2/STAT3 signaling to block enzalutamide-induced neuroendocrine differentiation [79].
Studies have demonstrated that plant-derived anticancer agents modulate the lncRNAs involved in prostate tumorigenesis like HOTAIR, H19, PCGEM1, DANCR, and lncRNAs-p21 [91, 92]. Aird et al. (2018) demonstrate that plant isoflavone genistein modulates the expression of HOTAIR in prostate cancer, which consequently modulates the activity of the PI3K/AKT signaling pathway [93]. Genistein induces apoptosis in prostate cancer cells by downregulating WNT and AKT signaling pathways, which might lead to decrease expression of HOTAIR in treated cells. In another study, Liu et al. (2019) have shown that polyphenol curcumin causes modulation of HOTAIR lncRNA in prostate cancer cells [94]. Similarly, resveratrol has been shown to inhibit AR signaling pathway in prostate cancer by modulating the expression of PCGEM1 and PRNCR1 lncRNAs [92]. Another study on resveratrol demonstrates a reduction of PCAT29 lncRNA expression in prostate cancer [95]. Liu et al. (2016) reported that 1,2,6-tri-O-galloyl-beta-D-glucopyranose has the potential to modulate lncRNA levels and could be a novel approach to treat prostate cancer [96].
The initiation and success of new genome editing tools including clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated (Cas) has promptly revolutionized the field of cancer biology. Although frequently used to engineer protein-coding genes, recent studies recommend that the technology can be applied to non-coding genes including lncRNAs, and to permanently correct in vivo genetic mutations. The technology can also benefit to identify genes involved in drug resistance to understand the mechanism of drug action and the efficacy of drug combinations in malignant cells. The use of this technology will help to uncover novel lncRNAs, to better annotate known lncRNAs, as well as to assess lncRNA localization, structure and function. Additional methodologies such as RNAi, antisense oligonucleotides, or small molecule inhibitors could be employed to better understand the biology of lncRNAs. Thus, given the roles of lncRNAs in various subtypes of prostate cancer, it will be important to recognize the therapeutic impact of targeting specific lncRNAs leading to a more efficient and broader therapeutic implication in prostate cancer.
Acknowledgements
PPK and MS acknowledge financial support from Indian Council of Medical Research (ICMR), India in the form of ICMR-Senior Research Fellowship. SK acknowledges University Grants Commission, India and Department of Science and Technology, India for providing financial support in the form of UGC-BSR Research Start-Up-Grant [No. F.30– 372/2017 (BSR)] and DST-SERB Grant [EEQ/2016/000350] and DST-FIST Departmental Grant. SK acknowledges Central University of Punjab, Bathinda, India for providing Research Seed Money Grant [GP-25]. AKS and KSP acknowledge CSIR-India and DBT-India funding agencies for providing financial assistance in the form of Senior and Junior Research Fellowships.
Ethical policy
Approval was taken from institutional ethical committee. The study was performed in accordance with the Declaration of Helsinki. Patients gave their informed consent for their participation.
Author contributions
SG and SK designed the study and drafted the manuscript. KSP, AKS and MS searched literature and compiled the data. KSP, AKS, and PPK prepared the figures and tables. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Funding
Efforts are supported by the Department of Defense Grants W81XWH-19-1-0720 and W81XWH-18-1-0618 and VA Merit Review 1I01BX002494 to SG.
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A: Global cancer statistics, 2012. CA: Cancer J Clin 2015, 65(2): 87-108.
- Kirby M, Hirst C, Crawford ED: Characterising the castration-resistant prostate cancer population: a systematic review. Int J Clin Pract 2011, 65(11): 1180-1192.
- Schweizer MT, Evan YY: Persistent androgen receptor addiction in castration-resistant prostate cancer. J Hematol Oncol 2015, 8: 1-4.
- Tiwari R, Manzar N, Ateeq B: Dynamics of Cellular Plasticity in Prostate Cancer Progression. Front Mol Biosci 2020, 7: 130.
- Wang HT, Yao YH, Li BG, Tang Y, Chang YT, Zhang J: Neuroendocrine Prostate Cancer (NEPC) Progressing from conventional prostatic adenocarcinoma: Factors associated with time to development of nepc and survival from NEPC Diagnosis-A systematic review and pooled analysis. J Clin Oncol 2014, 32(30): 3383-3390.
- Hauso O, Gustafsson BI, Kidd M, Waldum HL, Drozdov I, Chan AK, Modlin IM: Neuroendocrine tumor epidemiology: contrasting Norway and North America. Cancer 2008, 113(10): 2655-2664.
- Parimi V, Goyal R, Poropatich K, Yang XJ: Neuroendocrine differentiation of prostate cancer: a review. Am J Clin Exp Urol 2014, 2(4): 273-285.
- Lu NZ: The pharmacology and classification of the nuclear receptor superfamily: glucocorticoid, mineralocorticoid, progesterone, and androgen receptors. Pharmacol Rev 2006, 58(4): 782-797.
- Germain P, Staels B, Dacquet C, Spedding M, Laudet V: 2006. Overview of nomenclature of nuclear receptors. Pharmacol Rev 2006, 58(4): 685-704.
- Tan ME, Li J, Xu HE, Melcher K, Yong EL: Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacologica Sinica 2015, 36(1): 3-23.
- Messner EA, Steele TM, Tsamouri MM, Hejazi N, Gao AC, Mudryj M, Ghosh PM: The androgen receptor in prostate cancer: Effect of structure, ligands and spliced variants on therapy. Biomedicines 2020, 8(10): 422.
- Tucci M, Zichi C, Buttigliero C, Vignani F, Scagliotti GV, Di Maio M: Enzalutamide-resistant castration-resistant prostate cancer: challenges and solutions. OncoTargets Ther 2018, 11: 7353-7368.
- Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, Iversen P, Bhattacharya S, Carles J, Chowdhury S, Davis ID: Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med 2014, 371(5): 424-33.
- Chen R, Dong X, Gleave M: Molecular model for neuroendocrine prostate cancer progression. BJU Int 2018, 122(4): 560-570.
- Shorning BY, Dass MS, Smalley MJ, Pearson HB: The PI3K-AKT-mTOR Pathway and Prostate Cancer: At the Crossroads of AR, MAPK, and WNT Signaling. Int J Mol Sci 2020, 21(12): 1-47.
- Komiya A, Suzuki H, Imamoto T, Kamiya N, Nihei N, Naya Y, Ichikawa T, Fuse H: Neuroendocrine differentiation in the progression of prostate cancer. Int J Urol 2009, 16(1): 37-44.
- Terry S, Beltran H: The many faces of neuroendocrine differentiation in prostate cancer progression. Front Oncol 2014, 4: 60.
- Patel GK, Chugh N, Tripathi M: Neuroendocrine differentiation of prostate cancer—An intriguing example of tumor evolution at play. Cancers 2019, 11(10): 1405.
- Luo J, Wang K, Yeh S, Sun Y, Liang L, Xiao Y, Xu W, Niu Y, Cheng L, Maity SN, et al: LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat Commun 2019, 10(1): 2571.
- Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopolou E, Chakravarthi BVSK, Varambally S, et al: Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016, 22(3): 298-305.
- Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen C, Wongvipat J, Ku SY, Gao D, Cao Z, et al: SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Sci 2017, 355(6320): 84-88.
- Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, Labbe DP, Gomes EC, Wang J, Long HW, et al: Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017, 355(6320): 78-83.
- Dardenne E, Beltran H, Benelli M, Gayvert K, Berger A, Puca L, Cyrta J, Sboner A, Noorzad Z, MacDonald T, et al: N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell 2016, 30(4): 563-577.
- Lee JK, Phillips JW, Smith BA, Park JW, Stoyanova T, McCaffrey EF, Baertsch R, Sokolov A, Meyerowitz JG, Mathis C, et al: N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell 2016, 29(4): 536-547.
- Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, Wang Y, Sheikh KL, Terry S, Tagawa ST, et al: Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Disc 2011, 1(6): 487-495.
- Gopalakrishnan V: REST and RESTless. Future Neurol 2009, 4(3): 317-339.
- Lapuk AV, Wu C, Wyatt AW, McPherson A, McConeghy BJ, Brahmbhatt S, Mo F, Zoubeidi A, Anderson S, Bell RH, et al: From sequence to molecular pathology, and a mechanism driving the neuroendocrine phenotype in prostate cancer. J Pathol 2012, 227(3): 286-297.
- Zhang X, Coleman IM, Brown LG, True LD, Kollath L, Lucas JM, Lam HM, Dumpit R, Corey E, Chéry L, et al: SRRM4 expression and the loss of REST activity may promote the emergence of the neuroendocrine phenotype in castration-resistant prostate cancer. Clin Cancer Res 2015, 21(20): 4698-4708.
- Bishop JL, Thaper D, Vahid S, Davies A, Ketola K, Kuruma H, Jama R, Nip KM, Angeles A, Johnson F, et al: The master neural transcription factor BRN2 is an androgen receptor–suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov 2017, 7(1): 54-71.
- Qi J, Nakayama K, Cardiff RD, Borowsky AD, Kaul K, Williams R, Krajewski S, Mercola D, Carpenter PM, Bowtell D, et al: Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell 2010, 18(1): 23-38.
- Guo H, Ci X, Ahmed M, Hua JT, Soares F, Lin D, Puca L, Vosoughi A, Xue H, Li E, et al: ONECUT2 is a driver of neuroendocrine prostate cancer. Nature Commun 2019, 10(1): 278.
- Puca L, Vlachostergios PJ, Beltran H: Neuroendocrine differentiation in prostate cancer: emerging biology, models, and therapies. Cold Spring Harb Perspect Med 2019, 9(2): a030593.
- Rotinen M, You S, Yang J, Coetzee SG, Reis-Sobreiro M, Huang WC, Huang F, Pan X, Yáñez A, Hazelett DJ, et al: ONECUT2 is a targetable master regulator of lethal prostate cancer that suppresses the androgen axis. Nature Med 2018, 24(12): 1887-1898.
- Dicken H, Hensley PJ, Kyprianou N: Prostate tumor neuroendocrine differentiation via EMT: The road less traveled. As J Urol 2019, 6(1): 82-90.
- Huang X, Xiao R, Pan S, Yang X, Yuan W, Tu Z, Xu M, Zhu Y, Yin Q, Wu Y, et al: Uncovering the roles of long non-coding RNAs in cancer stem cells. J Hematol Oncol 2017, 10(1): 62.
- Schmitt AM, Chang HY: Long noncoding RNAs in cancer pathways. Cancer cell 2016, 29(4): 452-463.
- Ayub AL, Papaiz DD, da Silva Soares R, Jasiulionis MG: The Function of lncRNAs as Epigenetic Regulators in Non-Coding RNAs. Intech Open 2019, 7: 1-20.
- Fernandes JC, Acuña SM, Aoki JI, Floeter-Winter LM, Muxel SM: Long non-coding RNAs in the regulation of gene expression: physiology and disease. Non-coding RNA 2019, 5(1): 17.
- Jarroux J, Morillon A, Pinskaya M: History, discovery, and classification of lncRNAs. Adv Exp Med Biol 2017, 1008: 1-46.
- Rinn JL, Chang HY: Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012, 81: 145-166.
- Shih JW, Wang LY, Hung CL, Kung HJ, Hsieh CL: Non-Coding RNAs in Castration-Resistant Prostate Cancer: Regulation of Androgen Receptor Signaling and Cancer Metabolism. Int J Mol Sci 2015, 16(12): 28943-28978.
- Kim ST, Cheng Y, Hsu FC, Jin T, Kader AK, Zheng SL, Isaacs WB, Xu J, Sun J: Prostate cancer risk-associated variants reported from genome-wide association studies: meta-analysis and their contribution to genetic Variation. Prostate 2010, 70(16): 1729-1738.
- Jin G, Sun J, Isaacs SD, Wiley KE, Kim ST, Chu LW, Zhang Z, Zhao H, Zheng SL, Isaacs WB, et al: Human polymorphisms at long non-coding RNAs (lncRNAs) and association with prostate cancer risk. Carcinogen 2011, 32(11): 1655-1659.
- Grisanzio C, Freedman ML: Chromosome 8q24-Associated Cancers and MYC. Genes Cancer 2010, 1(6): 555-559.
- Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, Leongamornlert DA, Tymrakiewicz M, Jhavar S, Saunders E, et al: Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet 2009, 41(10): 1058-1060.
- Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al: lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 2013, 500(7464): 598-602.
- Hung CL, Wang LY, Yu YL, Chen HW, Srivastava S, Petrovics G, Kung HJ: A long noncoding RNA connects c-Myc to tumor metabolism. Proc Natl Acad Sci U S A. 2014, 111(52): 18697-18702.
- Işın M, Uysaler E, Özgür E, Köseoğlu H, Şanlı Ö, Yücel ÖB, Gezer U, Dalay N: Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front Genet 2015, 6: 1-5.
- Zhang A, Zhao JC, Kim J, Fong KW, Yang YA, Chakravarti D, Mo YY, Yu J: LncRNA HOTAIR Enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell Rep 2015, 13(1): 209-221.
- Xiang S, Zou P, Tang Q, Zheng F, Wu J, Chen Z, Hann SS: HOTAIR-mediated reciprocal regulation of EZH2 and DNMT1 contribute to polyphyllin I-inhibited growth of castration-resistant prostate cancer cells in vitro and in vivo. Biochim Biophys Acta Gen Subj 2018, 1862(3): 589-599.
- Misawa A, Takayama K, Urano T, Inoue S: Androgen-induced Long Noncoding RNA (lncRNA) SOCS2-AS1 Promotes Cell Growth and Inhibits Apoptosis in Prostate Cancer Cells. J Biol Chem 2016, 291(34): 17861-17880.
- Shang Z, Yu J, Sun L, Tian J, Zhu S, Zhang B, Dong Q, Jiang N, Flores-Morales A, Chang C, et al: LncRNA PCAT1 activates AKT and NF-κB signaling in castration-resistant prostate cancer by regulating the PHLPP/FKBP51/IKKα complex. Nucleic Acids Res 2019, 47(8): 4211-4225.
- Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S, Sahu A, Malik R, Wilder-Romans K, Navone N, et al: PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res 2014, 74(6): 1651-1660.
- Yang Z, Zhao S, Zhou X, Zhao H, Jiang X: PCAT-1: a pivotal oncogenic long non-coding RNA in human cancers. Biomed Pharmacother 2019, 1110: 493-499.
- Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, et al: The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun 2014, 5: 1-6.
- Ylipää A, Kivinummi K, Kohvakka A, Annala M, Latonen L, Scaravilli M, Kartasalo K, Leppänen SP, Karakurt S, Seppälä J, et al: Transcriptome Sequencing Reveals PCAT5 as a Novel ERG-Regulated Long Noncoding RNA in Prostate Cancer. Cancer Res 2015, 75(19): 4026-4031.
- Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, et al: The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet 2013, 45(11): 1392-1398.
- Liang X, Hu K, Li D, Wang Y, Liu M, Wang X, Zhu W, Wang X, Yang Z, Lu J: Identification of core genes and potential drugs for castration-resistant prostate cancer based on bioinformatics analysis. DNA Cell Biol 2020, 39(5): 836-847.
- Zhao M, Wang S, Li Q, Ji Q, Guo P, Liu X: MALAT1: A long non-coding RNA highly associated with human cancers. Oncol Lett 2018, 16(1): 19-26.
- Wang D, Ding L, Wang L, Zhao Y, Sun Z, Karnes RJ, Zhang J, Huang H: LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget 2015, 6(38): 41045-41055.
- Pickard MR, Mourtada-Maarabouni M, Williams GT: Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta 2013, 1832(10): 1613-1623.
- Yacqub-Usman K, Pickard MR, Williams, GT: Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. Prostate 2015, 75(7): 693-705.
- Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J, et al: lnc RNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J 2014, 281(16): 3766-3775.
- Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, Kwok TT: Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogen 2010, 31(3): 350-358.
- Dey BK, Pfeifer K, Dutta A: The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev 2014, 28(5): 491-501.
- Sun T, Du SY, Armenia J, Qu F, Fan J, Wang X, Fei T, Komura K, Liu SX, Lee GS, et al: Expression of lncRNA MIR222HG co-transcribed from the miR-221/222 gene promoter facilitates the development of castration-resistant prostate cancer. Oncogen 2018, 7(3): 30.
- Haddadi N, Lin Y, Travis G, Simpson AM, Nassif NT, McGowan EM: PTEN/PTENP1:‘Regulating the regulator of RTK-dependent PI3K/Akt signalling’, new targets for cancer therapy. Mol Cancer 2018, 17: 1-4.
- Ferraldeschi R, Rodrigues DN, Riisnaes R, Miranda S, Figueiredo I, Rescigno P, Ravi P, Pezaro C, Omlin A, Lorente D, et al: PTEN protein loss and clinical outcome from castration-resistant prostate cancer treated with abiraterone acetate. Europ Urol 2015, 67(4): 795-802.
- Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al: Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011, 147(2): 344-357.
- Ramnarine VR, Alshalalfa M, Mo F, Nabavi N, Erho N, Takhar M, Shukin R, Brahmbhatt S, Gawronski A, Kobelev M, et al: The long noncoding RNA landscape of neuroendocrine prostate cancer and its clinical implications. GigaSci 2018, 7(6): giy050.
- Duan Y, Li WX, Wang Y, Zhao Y, Shen J, Deng CJ, Li Q, Chen R, Liu X, Zhang YL: Integrated Analysis of lncRNAs and mRNAs Identifies a Potential Driver lncRNA FENDRR in Lung Cancer in Xuanwei, China. Nutr Cancer 2021, 73(6): 983-995.
- Zhang YQ, Chen X, Fu CL, Zhang W, Zhang DL, Pang C, Liu M, Wang JY: FENDRR reduces tumor invasiveness in prostate cancer PC-3 cells by targeting CSNK1E. Eur Rev Med Pharmacol Sci 2019, 23(17):7 327-7337.
- Mirza S, Katafiasz BJ, Kumar R, Wang J, Mohibi S, Jain S, Gurumurthy CB, Pandita TK, Dave BJ, Band H, et al: Alteration/deficiency in activation-3 (Ada3) plays a critical role in maintaining genomic stability. Cell Cycle 2012, 11(22): 4266-4274.
- Klomp MJ, Dalm SU, de Jong M, Feelders RA, Hofland J, Hofland LJ: Epigenetic regulation of somatostatin and somatostatin receptors in neuroendocrine tumors and other types of cancer. Rev Endocr Metab Disord 2020, 22(3): 495-510.
- Mather RL, Venalainen E, Pucci P, Lin D, Xue H, Wang Y, Crea F: The long non-coding RNA NEAR1 promotes neuroendocrine prostate cancer cell proliferation and survival. 3rd Int Cancer Symp Cancer Res Center 2017, 25-27.
- Raveh E, Matouk IJ, Gilon M, Hochberg A: The H19 Long non-coding RNA in cancer initiation, progression and metastasis–a proposed unifying theory. Mol Cancer 2015, 14: 184.
- Sun SC, Zhao HR, Liu BL, Wang YQ, Liu Y, Zhao, Shi ZD: [Expression of long non-coding RNA H19 in prostate cancer and its effect on the proliferation and glycometabolism of human prostate cancer cells]. Natl J Androl 2017, 23(2): 120-124.
- Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA, et al: LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell 2014, 54(5): 777-790.
- Luo J, Wang K, Yeh S, Sun Y, Liang L, Xiao Y, Xu W, Niu Y, Cheng L, Maity SN, et al: LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nature Commun 2019, 10(1): 2571.
- Chang YT, Lin TP, Tang JT, Campbell M, Luo YL, Lu SY, Yang CP, Cheng TY, Chang CH, Liu TT, et al: HOTAIR is a REST-regulated lncRNA that promotes neuroendocrine differentiation in castration resistant prostate cancer. Cancer Lett 2018, 433: 43-52.
- Li L, Dang Q, Xie H, Yang Z, He D, Liang L, Song W, Yeh S, Chang C: Infiltrating mast cells enhance prostate cancer invasion via altering LncRNA-HOTAIR/PRC2-androgen receptor (AR)-MMP9 signals and increased stem/progenitor cell population. Oncotarget 2015, 6(16): 14179-14190.
- Li Y, Luo H, Xiao N, Duan J, Wang Z, Wang S: Long noncoding RNA SChLAP1 accelerates the proliferation and metastasis of prostate cancer via targeting miR-198 and promoting the MAPK1 pathway. Oncol Res Featur Preclin Clin Cancer Ther 2018, 26(1): 131-143.
- Chang J, Xu W, Du X, Hou J: MALAT1 silencing suppresses prostate cancer progression by upregulating miR-1 and downregulating KRAS. OncoTar Ther 2018, 11: 3461-3473.
- Ling Z, Wang X, Tao T, Zhang L, Guan H, You Z, Lu K, Zhang G, Chen S, Wu J, et al: Involvement of aberrantly activated HOTAIR/EZH2/miR-193a feedback loop in progression of prostate cancer. J Exp Clin Cancer Res 2017, 36(1): 159.
- Crea F, Venalainen E, Ci X, Cheng H, Pikor L, Parolia A, Xue H, Nur Saidy NR, Lin D, Lam W, et al: The role of epigenetics and long noncoding RNA MIAT in neuroendocrine prostate cancer. Epigenom 2016, 8(5): 721-731.
- Ozgur E, Gezer U: Investigation of lncRNA H19 in prostate cancer cells and secreted exosomes upon androgen stimulation or androgen receptor blockage. Bratisl Lek Listy 2020, 121(5): 362-365.
- Yacqub-Usman K, Pickard MR, Williams GT: Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. The prostate 2015, 75(7): 693-705.
- Xiang S, Zou P, Wu J, Zheng F, Tang Q, Zhou J, Hann SS: Crosstalk of NF-κB/P65 and LncRNA HOTAIR-Mediated Repression of MUC1 Expression Contribute to Synergistic Inhibition of Castration-Resistant Prostate Cancer by Polyphyllin 1–Enzalutamide Combination Treatment. Cell Physiol Biochem 2018, 47(2): 759-773.
- Zhao HF, Zhang ZC, Shi BK, Jiang XZ: DANCR sponges miR-135a to regulate paclitaxel sensitivity in prostate cancer. Eur Rev Med Pharmacol Sci 2019, 23(16): 6849-6857.
- Karantanos T, Corn P G, Thompson TC: Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene 2013, 32(49): 5501-5511.
- Li CH, Chen Y: Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol 2013, 45(8): 1895-1910.
- Saghafi T, Taheri RA, Parkkila S, Zolfaghari Emameh R: Phytochemicals as modulators of long non-coding RNAs and inhibitors of cancer-related carbonic anhydrases. Int J Mol Sci 2019, 20(12): 2939.
- Aird J, Baird AM, Lim MC, McDermott R, Finn SP, Gray SG: Carcinogenesis in prostate cancer: the role of long non-coding RNAs. Non-coding RNA Res 2018, 3(1): 29-38.
- Liu Y, Sun H, Makabel B, Cui Q, Li J, Su C, Ashby Jr CR, Chen Z, Zhang J: The targeting of noncoding RNAs by curcumin: Facts and hopes for cancer therapy. Oncol Rep 2019, 42(1): 20-34.
- Al Aameri RF, Sheth S, Alanisi EM, Borse V, Mukherjea D, Rybak LP, Ramkumar V: Tonic suppression of PCAT29 by the IL-6 signaling pathway in prostate cancer: Reversal by resveratrol. PLoS One 2017, 12(5): e0177198.
- Liu D, Yu X, Wang S, Dai E, Jiang L, Wang J, Yang Q, Yang F, Zhou S, Jiang W: The gain and loss of long noncoding RNA associated-competing endogenous RNAs in prostate cancer. Oncotarget 2016, 7(35): 57228-57238.
Annals of urologic oncology
p-ISSN: 2617-7765, e-ISSN: 2617-7773
 Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript