Review Article | Open Access
Research Progress of Abnormal DNA Methylation in the Development, Diagnosis, and Treatment of Prostate Cancer
Zhen Ren1, Ming Yu1, Yi Xia2
1Department of Ultrasound, Xijing Hospital, Fourth Military Medical University, Xi'an 710032, China.
2Department of Neurosurgery, Tangdu Hospital, Fourth Military Medical University, Xi'an 710038, China.
Correspondence: Yi Xia (Department of Neurosurgery, Tangdu Hospital, Fourth Military Medical University, Xinshi Road, Xi'an, Shaanxi Province, P.R. China, 710038; Email: shylock_xy@163.com) and Ming Yu (Department of Ultrasound, Xijing Hospital, Fourth Military Medical University, Changle West Road, Xi'an, Shaanxi Province, P.R. China, 710000; Email: yumingfmmu@126.com).
Annals of Urologic Oncology 2023, 6(3): 96-103. https://doi.org/10.32948/auo.2023.09.30
Received: 12 Sep 2023 | Accepted: 30 Sep 2023 | Published online: 03 Oct 2023
Key words DNA methylation, epigenetics, prostate cancer, diagnosis, treatment
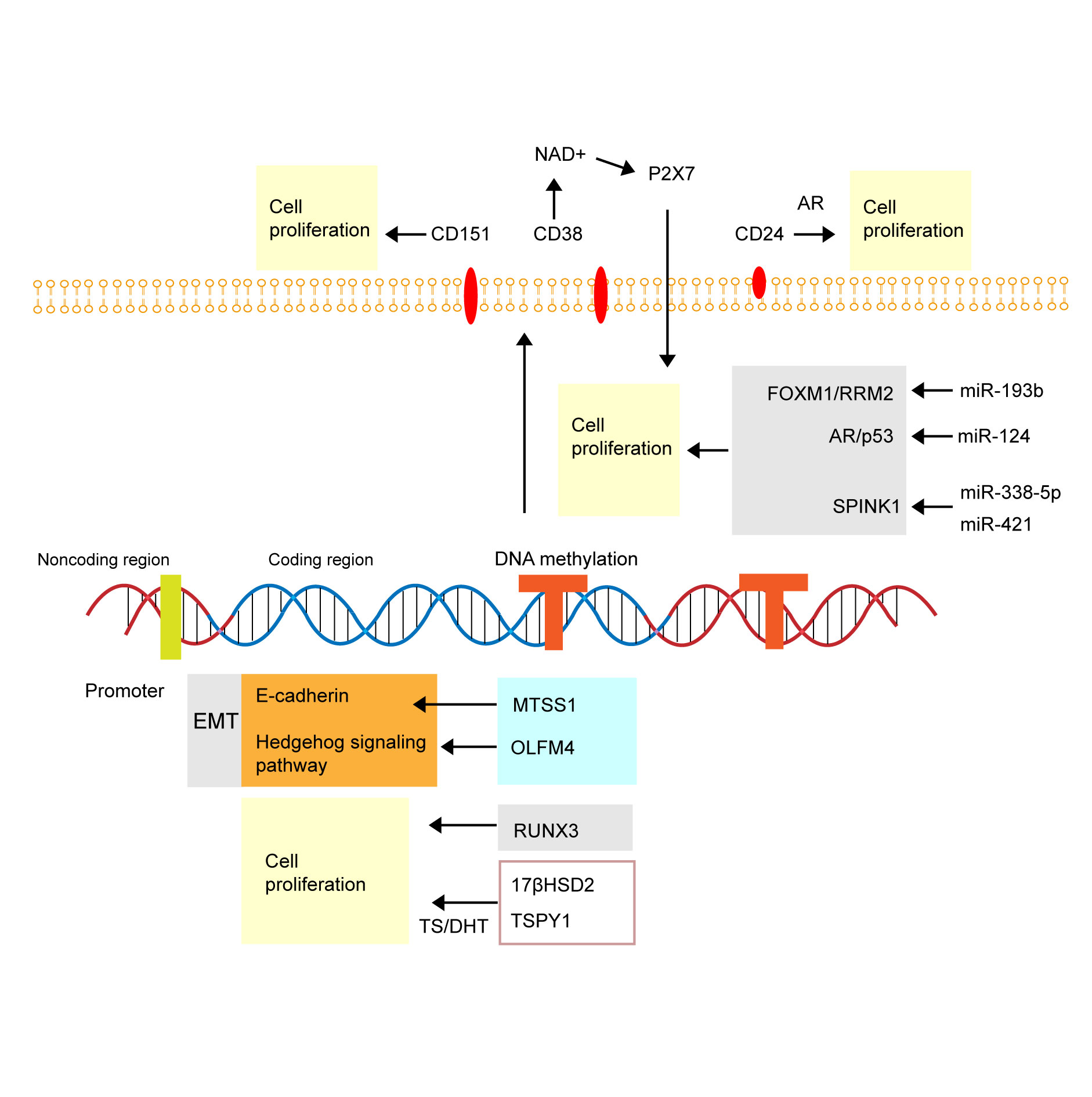 Figure 1. Graph of molecular mechanism of abnormal DNA methylation in prostate cancer. EMT, Epithelial-mesenchymal transition; MTSS1, metastasis suppressor 1; OLFM4, metastasis suppressor 1; RUNX3, RUNt-related transcription factor 3; 17βHSD2, type 2 17β-hydroxysteroidal solid ol dehydrogenase; TSPY1, testis-specific protein Y-encoded 1; AR, androgen receptor.
Figure 1. Graph of molecular mechanism of abnormal DNA methylation in prostate cancer. EMT, Epithelial-mesenchymal transition; MTSS1, metastasis suppressor 1; OLFM4, metastasis suppressor 1; RUNX3, RUNt-related transcription factor 3; 17βHSD2, type 2 17β-hydroxysteroidal solid ol dehydrogenase; TSPY1, testis-specific protein Y-encoded 1; AR, androgen receptor.
Early screening and diagnosis of PCa in clinic mainly rely on PSA detection.
However, the PSA level is affected by various factors, and the specificity and sensitivity are not optimal. DNA methylation is highly specific and stable in PCa, so it has its unique advantages in tumor diagnosis. Yao et al. [25] selected a methylation site on Y chromosome (cg05163709) as a potential biomarker for PCa diagnosis through cancer tissues and adjacent normal tissues of 66 PCa patients, and its sensitivity and specificity were 94.6% and 78.3% respectively. ROC analysis showed that the diagnostic efficiency of cg05163709 (AUC = 0.915) was significantly higher than that of PSA test (AUC = 0.769). Li et al. [26] identified 359 hypermethylation sites, 3 435 hypomethylation sites, 483 up-regulated genes and 1 341 down-regulated genes in PCa using a public database, and selected 17 hypermethylation sites (covering 13 genes) that could highly distinguish PCa from normal tissues. The prediction accuracy of PCa diagnosis was 88%-94%, respectively. These markers provided important clues for PCa diagnosis and prognosis assessment. Lan et al. [27] detected the methylation pattern of DACT-2 in serum of 64 PCa patients, 22 patients with benign prostatic hyperplasia (BPH) and 47 healthy subjects by methylation-specific PCR, and the results showed that DACT-2 was present in PCa patients. The promoter methylation level was significantly higher than that of BPH patients and healthy subjects, and the methylation rate of DACT-2 was 0.745. The sensitivity was 81.8% and the specificity was 75.0%. Meanwhile, the ROC curve results showed that DACT-2 was better than PSA in the diagnosis of PCa and was a potential biomarker for PCa diagnosis.
The combined methylation test of multiple genes is helpful to improve the sensitivity and specificity of PCa diagnosis. Brait et al. [28] found that the sensitivity and specificity of MCAM hypermethylation for the diagnosis of PCa was 66% and 73%, and when MCAM was combined with ERα and ERβ, the sensitivity was effectively increased to 75%, but the specificity was not significantly reduced. Haldrup et al. [25] analyzed the methylation of serum samples from 27 patients with PCa and 10 patients with BPH, and finally identified three hypermethylated genes (ST6GALNAC3 / CCDC181 / HAPLN3 genes) for the construction of PCa diagnostic model, with high sensitivity. The sensitivity was 67% and the specificity was 100%. Constancio et al. [29] found that the accuracy, sensitivity and specificity of the combined detection of FOXA1, GSTP1, HOXD3, RARβ2, RASSF1A, SEPT9 and SOX17 for PCa diagnosis was 72%. These studies indicate that polygene methylation detection is expected to become a new idea for PCa diagnosis. DNA methylation test combined with serum PSA also has a common role in PCa diagnosis. A Mexican cohort study [30] found that methylation tests for GSTP1 and RASSF1A genes had a positive predictive value of 73% and a negative predictive value of 59.6% for PCa diagnosis. When combined with serum PSA, the positive and negative predictive values increased to 81% and 66%, respectively. Reis et al. [31] compared serum GADD45 for a methylation levels in 22 PCa patients and 22 control patients, and found that the sensitivity and specificity of GADD45 methylation in serum samples were 38% and 98%. However, as a PCa biomarker combination consisting of PSA, circulating cell free DNA (cfDNA) level and GADD45a methylation was established, although the specificity was slightly decreased (87.5%), the diagnostic sensitivity was significantly improved (94.1%). ROC analysis showed that the combination had good diagnostic efficacy for PCa (AUC = 0.937). Figure 2 shows that cfDNA can be released in the bloodstream from tumor sites in prostate cancer patients.
Taken together, these studies suggest that abnormal DNA methylation may be a potential biomarker for early diagnosis of PCa. At the same time, detection of methylation sites based on body fluids or tissues has the characteristics of non-invasive, economical and convenient, which sets a new direction of cancer diagnosis in the future. However, whether DNA methylation is consistent in blood and tumor tissues needs further research.
Application of DNA methylation in PCa prognostic assessment
DNA methylation is not only involved in the initiation and development of PCa, but also closely related to poor prognosis of PCa. It can be used as an important marker for the assessment of PCa prognosis. In a prospective study, Wang et al. [32] found that the methylation pattern of serum cadherin-13 (CDH13) gene not only significantly correlated with Gleason score, tumor stage and PSA level, but also strongly correlated with survival outcomes and relative mortality risk (HR = 6.132, 95%CI: 3.160-12.187). Gao et al. [33] found that the promoter methylation level of CRMP4 gene could be used as a factor.
The sensitivity and specificity of this method was more than 90% and served as independent predictors of prognosis in PCa patients. The sensitivity of this method was significantly better than that of MRI in diagnosing lymph node metastasis of PCa (P < 0.001). Pellacani et al. [34] utilized a detection method based on 17 methylation markers for stratification of PCa prognosis based on whole-genome DNA methylation analysis in PCa tissues and normal tissues, and the diagnostic accuracy rate of this method was as high as 92%. Zhang et al. [35] divided PCa patients into hypermethylated and hypomethylated types based on DNA methylation patterns at CpG sites. The two subtypes showed significant differences in the epigenome, genome, transcriptome, disease state, immune cell infiltration and function, among which 8 were associated with high-risk subtypes. The PCa prognostic evaluation model constructed by AURKA, DLGAP5, FOXD1, KIF4A, MELK, MYBL2, SPAG5 and TPX2 showed that a higher risk score was associated with poor prognosis in PCa (P < 0.05). The validity of the model was 66%-84% in both internal and external validation sets. In conclusion, high specificitysensitivity and diagnostic accuracy of DNA methylation detection in PCa is applicable for prognosis assessment. In addition, a large number of studies have found that the level of DNA methylation was closely related to the biochemical recurrence of PCa. Goltz et al. [36] found that CXCL12 methylation significantly correlated with Gleason grade and biochemical relape-free survival after radical prostatectomy, and could be used as an active indicator of postoperative biochemical relapse in PCa patients.
Norgaard et al. [37] found that MEIS2 gene was significantly hypermethylated in PCa, and in 3 independent radical anterior adenectomy cohorts (700 patients in total), The expression level of MEIS2 significantly correlated with postoperative biochemical recurrence (P.0084, 0.001 and 0.01,). Holmes et al. [38] found that promoter methylation of PITX3 could effectively predict biochemical recurrence of PCa (training set: HR = 1.83, 95%CI: 1.07 ~ 3.11, P = 0.027; Validation set: HR = 2.56, 95%CI: 1.44 ~ 4.54, P = 0.001), and the combination of PITX2 detection could also perform risk stratification in PCa patients to better assess the risk of biochemical recurrence. In conclusion, detection of methylation markers can predict the risk of biochemical recurrence in PCa patients after surgery, which is of great significance for the prognosis and PCa monitoring.
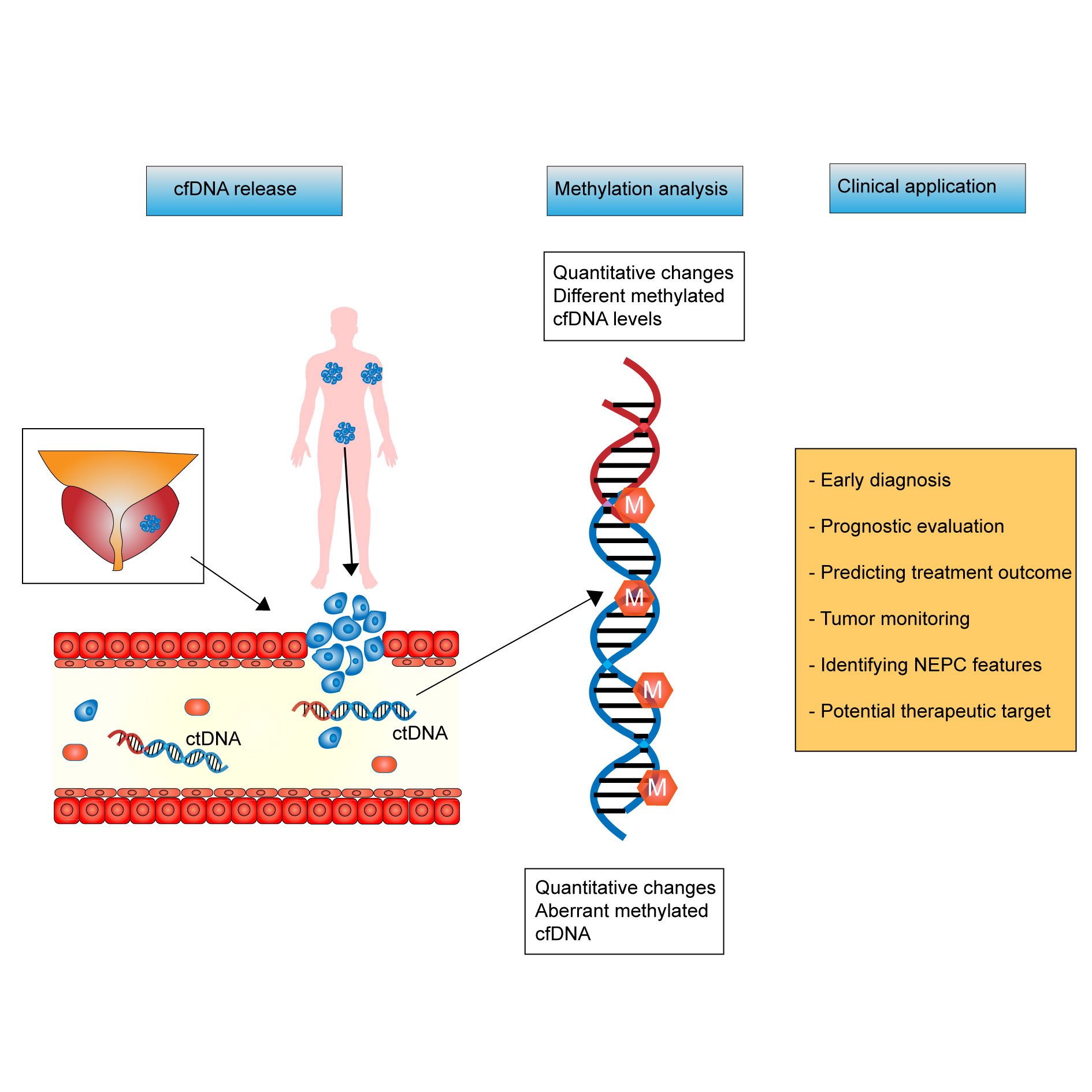 Figure 2. Potential clinical application of cell free DNA and abnormal DNA methylation in prostate cancer. Circulating cell free DNA (cfDNA) can be released in the bloodstream from tumor sites in prostate cancer patients. cfDNA form the bloodstream in prostate cancer patients can undergo both quantitative (e.g., changes in levels of methylated DNA) and qualitative (e.g., specific patterns of aberrant methylation) changes. Its evaluation of the methylation in cfDNA can help manage prostate cancer patients resulting in early diagnosis, a prognostic and predictive evaluation, a better tumor monitoring, and may aid in the identification of resistance such as neuroendocrine prostate cancer (NEPC).
Figure 2. Potential clinical application of cell free DNA and abnormal DNA methylation in prostate cancer. Circulating cell free DNA (cfDNA) can be released in the bloodstream from tumor sites in prostate cancer patients. cfDNA form the bloodstream in prostate cancer patients can undergo both quantitative (e.g., changes in levels of methylated DNA) and qualitative (e.g., specific patterns of aberrant methylation) changes. Its evaluation of the methylation in cfDNA can help manage prostate cancer patients resulting in early diagnosis, a prognostic and predictive evaluation, a better tumor monitoring, and may aid in the identification of resistance such as neuroendocrine prostate cancer (NEPC).
At present, there are few clinical trials on DNMTi for PCa, and most of them are phase I and II clinical trials. Sonpavde et al. [44] recruited 36 patients with castration-resistant Pca for the phase II trial of azacytidine combined with androgen blocking agent, and results showed that the PSA doubling time of 19 patients was greater than 3 months, and the overall median PSA doubling time of patients was significantly longer than baseline (P < 0. 01), which is related to the reduction of DNALINE-1 methylation in plasma by azacitidine. A small Phase II clinical trial [45] involving 14 mCRPC patients using three intravenous doses of decitabine 75 mg/m2 every 8 hours, repeated every 5 to 8 weeks, showed that the patients were well tolerated, but only 2 patients had stable disease and disease progression takes longer than 10 weeks. Another phase I/ II study [46] used azacytidine plus docetaxel plus prednisone to treat mCRPC patients, with 15 and 7 patients enrolled in phase I and II trials, ultimately observed a complete response in 1 patient and a partial response in 2 patients out of 10 evaluable patients. There were five patients with stable disease. These clinical trials indicate that the safety and clinical efficacy of DNMTi need to be further studied and improved, and that targeted therapy for abnormal DNA methylation sites should be developed to achieve better results of PCa treatment.
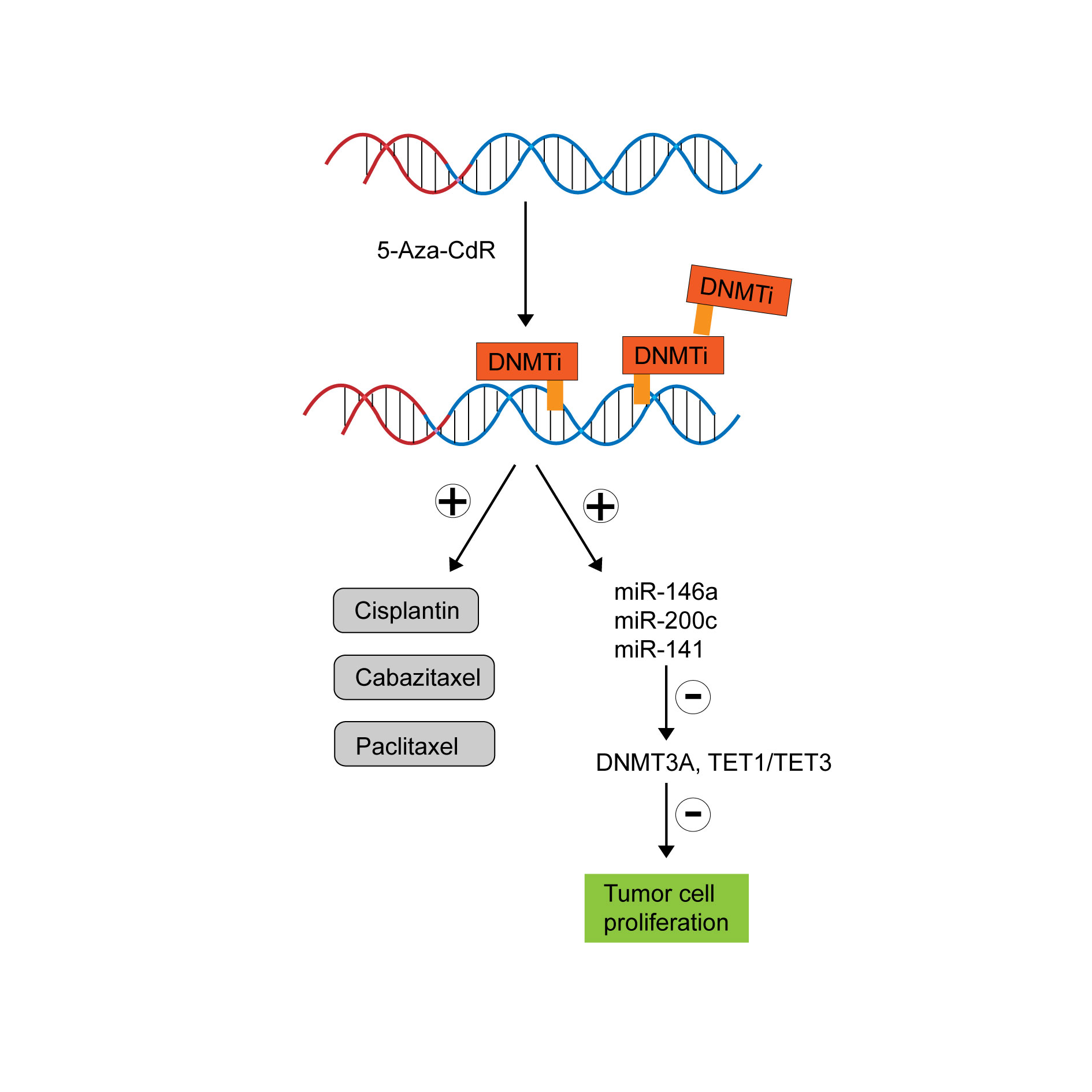 Figure 3. Graph of DNMTi (5 - Aza - CdR) for therapy of prostate cancer.
Figure 3. Graph of DNMTi (5 - Aza - CdR) for therapy of prostate cancer.
None.
Ethical policy
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study. Approval from institutional ethical committee was taken.
Availability of data and materials
All data generated or analysed during this study are included in this publication.
Author contributions
ZR: Conception, design of study and manuscript preparation; MY: Data collection and analysis; YX: Approval for the final version of the manuscript and funding supports.
Competing interests
The authors have no competing interest.
Funding
This article is funded by Shaanxi Province Project (2018JQ8075).
- Siegel RL, Miller KD, Fuchs HE, Jemal A: Cancer statistics, 2022. CA Cancer J Clin 2022, 72(1): 7-33.
- Liu Z, Meng X, Zhang H, Li Z, Liu J, Sun K, Meng Y, Dai W, Xie P, Ding Y et al: Predicting distant metastasis and chemotherapy benefit in locally advanced rectal cancer. Nat Commun 2020, 11(1): 4308.
- Shou J, Zhang Q, Wang S, Zhang D: The prognosis of different distant metastases pattern in prostate cancer: A population based retrospective study. Prostate 2018, 78(7): 491-497.
- Chang T, Lian Z, Ma S, Liang Z, Ma X, Wen X, Wang Y, Liu R: Combination with vorinostat enhances the antitumor activity of cisplatin in castration-resistant prostate cancer by inhibiting DNA damage repair pathway and detoxification of GSH. Prostate 2023, 83(5): 470-486.
- Rocha SM, Sousa I, Gomes IM, Arinto P, Costa-Pinheiro P, Coutinho E, Santos CR, Jeronimo C, Lemos MC, Passarinha LA et al: Promoter Demethylation Upregulates STEAP1 Gene Expression in Human Prostate Cancer: In Vitro and In Silico Analysis. Life (Basel) 2021, 11(11).
- Kim YJ, Yoon HY, Kim SK, Kim YW, Kim EJ, Kim IY, Kim WJ: EFEMP1 as a novel DNA methylation marker for prostate cancer: array-based DNA methylation and expression profiling. Clin Cancer Res 2011, 17(13): 4523-4530.
- Chattopadhyaya S, Ghosal S: DNA methylation: a saga of genome maintenance in hematological perspective. Hum Cell 2022, 35(2): 448-461.
- Kulis M, Esteller M: DNA methylation and cancer. Adv Genet 2010, 70: 27-56.
- Aref-Eshghi E, Schenkel LC, Ainsworth P, Lin H, Rodenhiser DI, Cutz JC, Sadikovic B: Genomic DNA Methylation-Derived Algorithm Enables Accurate Detection of Malignant Prostate Tissues. Front Oncol 2018, 8: 100.
- Xu N, Wu YP, Ke ZB, Liang YC, Cai H, Su WT, Tao X, Chen SH, Zheng QS, Wei Y et al: Identification of key DNA methylation-driven genes in prostate adenocarcinoma: an integrative analysis of TCGA methylation data. J Transl Med 2019, 17(1): 311.
- Zhao SG, Chen WS, Li H, Foye A, Zhang M, Sjostrom M, Aggarwal R, Playdle D, Liao A, Alumkal JJ et al: The DNA methylation landscape of advanced prostate cancer. Nat Genet 2020, 52(8): 778-789.
- Yan W, Guo Y, Xu F, Saxena D, Li X: Identification of Differentially Methylated Regions Associated with a Knockout of SUV39H1 in Prostate Cancer Cells. Genes (Basel) 2020, 11(10).
- Yin L, Zhu Z, Huang L, Luo X, Li Y, Xiao C, Yang J, Wang J, Zou Q, Tao L et al: DNA repair- and nucleotide metabolism-related genes exhibit differential CHG methylation patterns in natural and synthetic polyploids (Brassica napus L.). Hortic Res 2021, 8(1): 142.
- Yang X, Wang S, Reheman A: Regulation of RUNX3 Expression by DNA Methylation in Prostate Cancer. Cancer Manag Res 2020, 12: 6411-6420.
- Chen J, Huang L, Zhu Q, Wang Z, Tang Z: MTSS1 hypermethylation is associated with prostate cancer progression. J Cell Physiol 2020, 235(3): 2687-2697.
- Li H, Kim C, Liu W, Zhu J, Chin K, Rodriguez-Canales J, Rodgers GP: Olfactomedin 4 downregulation is associated with tumor initiation, growth and progression in human prostate cancer. Int J Cancer 2020, 146(5): 1346-1358.
- Gao X, Dai C, Huang S, Tang J, Chen G, Li J, Zhu Z, Zhu X, Zhou S, Gao Y et al: Functional Silencing of HSD17B2 in Prostate Cancer Promotes Disease Progression. Clin Cancer Res 2019, 25(4): 1291-1301.
- Leng X, Liu M, Tao D, Yang B, Zhang Y, He T, Xie S, Wang Z, Liu Y, Yang Y: Epigenetic modification-dependent androgen receptor occupancy facilitates the ectopic TSPY1 expression in prostate cancer cells. Cancer Sci 2021, 112(2): 691-702.
- Mazzu YZ, Yoshikawa Y, Nandakumar S, Chakraborty G, Armenia J, Jehane LE, Lee GM, Kantoff PW: Methylation-associated miR-193b silencing activates master drivers of aggressive prostate cancer. Mol Oncol 2019, 13(9): 1944-1958.
- Shi J, Hu J, Zhou Q, Du Y, Jiang C: PEpiD: a prostate epigenetic database in mammals. PLoS One 2013, 8(5): e64289.
- Bhatia V, Yadav A, Tiwari R, Nigam S, Goel S, Carskadon S, Gupta N, Goel A, Palanisamy N, Ateeq B: Epigenetic Silencing of miRNA-338-5p and miRNA-421 Drives SPINK1-Positive Prostate Cancer. Clin Cancer Res 2019, 25(9): 2755-2768.
- Mottahedeh J, Haffner MC, Grogan TR, Hashimoto T, Crowell PD, Beltran H, Sboner A, Bareja R, Esopi D, Isaacs WB et al: CD38 is methylated in prostate cancer and regulates extracellular NAD(). Cancer Metab 2018, 6: 13.
- Han R, Hensley PJ, Li J, Zhang Y, Stark TW, Heller A, Qian H, Shi J, Liu Z, Huang JA et al: Integrin-associated CD151 is a suppressor of prostate cancer progression. Am J Transl Res 2020, 12(4): 1428-1442.
- Tolkach Y, Zarbl R, Bauer S, Ritter M, Ellinger J, Hauser S, Huser L, Klauck SM, Altevogt P, Sultmann H et al: DNA Promoter Methylation and ERG Regulate the Expression of CD24 in Prostate Cancer. Am J Pathol 2021, 191(4): 618-630.
- Yao L, Ren S, Zhang M, Du F, Zhu Y, Yu H, Zhang C, Li X, Yang C, Liu H et al: Identification of specific DNA methylation sites on the Y-chromosome as biomarker in prostate cancer. Oncotarget 2015, 6(38): 40611-40621.
- Li Y, Meng L, Shi T, Ren J, Deng Q: Diagnosis and prognosis potential of four gene promoter hypermethylation in prostate cancer. Cell Biol Int 2021, 45(1): 117-126.
- Lan T, Yin L, Zhang H, Gu B, Ma P, Li S, Li H: Diagnostic value of DACT-2 methylation in serum of prostate cancer patients. Ann Palliat Med 2021, 10(3): 2421-2428.
- Liu JW, Nagpal JK, Jeronimo C, Lee JE, Henrique R, Kim MS, Ostrow KL, Yamashita K, van Criekinge V, Wu G et al: Hypermethylation of MCAM gene is associated with advanced tumor stage in prostate cancer. Prostate 2008, 68(4): 418-426.
- Haldrup C, Pedersen AL, Ogaard N, Strand SH, Hoyer S, Borre M, Orntoft TF, Sorensen KD: Biomarker potential of ST6GALNAC3 and ZNF660 promoter hypermethylation in prostate cancer tissue and liquid biopsies. Mol Oncol 2018, 12(4): 545-560.
- Sanchez BE, Aguayo A, Martinez B, Rodriguez F, Marmolejo M, Svyryd Y, Luna L, Munoz LA, Jimenez MA, Sotomayor M et al: Using Genetic and Epigenetic Markers to Improve Differential Diagnosis of Prostate Cancer and Benign Prostatic Hyperplasia by Noninvasive Methods in Mexican Patients. Clin Genitourin Cancer 2018, 16(4): e867-e877.
- Reis IM, Ramachandran K, Speer C, Gordian E, Singal R: Serum GADD45a methylation is a useful biomarker to distinguish benign vs malignant prostate disease. Br J Cancer 2015, 113(3): 460-468.
- Wang L, Lin YL, Li B, Wang YZ, Li WP, Ma JG: Aberrant promoter methylation of the cadherin 13 gene in serum and its relationship with clinicopathological features of prostate cancer. J Int Med Res 2014, 42(5): 1085-1092.
- Gao X, Mao YH, Xiao C, Li K, Liu W, Li LY, Pang J: Calpain-2 triggers prostate cancer metastasis via enhancing CRMP4 promoter methylation through NF-kappaB/DNMT1 signaling pathway. Prostate 2018, 78(9): 682-690.
- Pellacani D, Droop AP, Frame FM, Simms MS, Mann VM, Collins AT, Eaves CJ, Maitland NJ: Phenotype-independent DNA methylation changes in prostate cancer. Br J Cancer 2018, 119(9): 1133-1143.
- Zhang X, Wu M, Xiao H, Lee MT, Levin L, Leung YK, Ho SM: Methylation of a single intronic CpG mediates expression silencing of the PMP24 gene in prostate cancer. Prostate 2010, 70(7): 765-776.
- Goltz D, Holmes EE, Gevensleben H, Sailer V, Dietrich J, Jung M, Rohler M, Meller S, Ellinger J, Kristiansen G et al: CXCL12 promoter methylation and PD-L1 expression as prognostic biomarkers in prostate cancer patients. Oncotarget 2016, 7(33): 53309-53320.
- Norgaard M, Haldrup C, Bjerre MT, Hoyer S, Ulhoi B, Borre M, Sorensen KD: Epigenetic silencing of MEIS2 in prostate cancer recurrence. Clin Epigenetics 2019, 11(1): 147.
- Holmes EE, Goltz D, Sailer V, Jung M, Meller S, Uhl B, Dietrich J, Rohler M, Ellinger J, Kristiansen G et al: PITX3 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients after radical prostatectomy. Clin Epigenetics 2016, 8: 104.
- Wang X, Gao H, Ren L, Gu J, Zhang Y, Zhang Y: Demethylation of the miR-146a promoter by 5-Aza-2'-deoxycytidine correlates with delayed progression of castration-resistant prostate cancer. BMC Cancer 2014, 14: 308.
- Perry AS, O'Hurley G, Raheem OA, Brennan K, Wong S, O'Grady A, Kennedy AM, Marignol L, Murphy TM, Sullivan L et al: Gene expression and epigenetic discovery screen reveal methylation of SFRP2 in prostate cancer. Int J Cancer 2013, 132(8): 1771-1780.
- Fang XM, Jiang ZH, Yao N, Ding XW, Peng JP, Zheng S: [Effects of 5-Aza-2'-deoxycytidine on the carcinogenesis of colorectal cancer in mouse and the in vivo expression of p16/CDKN(2) mRNA]. Zhonghua Yi Xue Za Zhi 2011, 91(33): 2362-2365.
- Ramachandran K, Speer C, Nathanson L, Claros M, Singal R: Role of DNA Methylation in Cabazitaxel Resistance in Prostate Cancer. Anticancer Res 2016, 36(1): 161-168.
- Fan L, Li H, Zhang Y: LINC00908 negatively regulates microRNA-483-5p to increase TSPYL5 expression and inhibit the development of prostate cancer. Cancer Cell Int 2020, 20: 10.
- Sonpavde G, Aparicio AM, Zhan F, North B, Delaune R, Garbo LE, Rousey SR, Weinstein RE, Xiao L, Boehm KA et al: Azacitidine favorably modulates PSA kinetics correlating with plasma DNA LINE-1 hypomethylation in men with chemonaive castration-resistant prostate cancer. Urol Oncol 2011, 29(6): 682-689.
- Thibault A, Figg WD, Bergan RC, Lush RM, Myers CE, Tompkins A, Reed E, Samid D: A phase II study of 5-aza-2'deoxycytidine (decitabine) in hormone independent metastatic (D2) prostate cancer. Tumori 1998, 84(1): 87-89.
- Singal R, Ramachandran K, Gordian E, Quintero C, Zhao W, Reis IM: Phase I/II study of azacitidine, docetaxel, and prednisone in patients with metastatic castration-resistant prostate cancer previously treated with docetaxel-based therapy. Clin Genitourin Cancer 2015, 13(1): 22-31.
Annals of urologic oncology
p-ISSN: 2617-7765, e-ISSN: 2617-7773
 Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript