Review Article | Open Access
The Roles of Ferroptosis-related Long Non-coding RNAs in Urologic Cancers
Wenchao Xie1, Jie Gu2, Zhenqian Qin1, Yimin Xie11Department of Urology, Affiliated Hospital of Jiangsu University-Yixing Hospital, Yixing, Jiangsu, 214200, China.
2School of Life Sciences, Jiangsu University, Zhenjiang, Jiangsu, 212013, China.
Correspondence: Yimin Xie (Department of Urology, Affiliated Hospital of Jiangsu University-Yixing Hospital, Yixing, Jiangsu, 214200, China; Email: 2221513084@stmail.ujs.edu.cn).
Annals of Urologic Oncology 2023, 6(1): 3-9. https://doi.org/10.32948/auo.2023.01.20
Received: 11 Jan 2023 | Accepted: 19 Jan 2023 | Published online: 30 Jan 2023
Ferroptosis is a type of programmed cell death that has been recent topic of interest in cancer therapy. Growing evidence indicates that long noncoding RNAs (lncRNAs) are involved in ferroptosis and associated with the incidence and progression of cancer. However, the relationship between lncRNA and ferroptosis in urologic cancers has not been fully elucidated. In this review, we summarize ferroptosis-related lncRNAs (frlncRNAs) in urologic cancers. Studies indicate that frlncRNAs are associated with cancer metabolism, tumor microenvironment, and immune cell infiltration. In addition, frlncRNAs could regulate ferroptosis related genes both at the mRNA and protein level. Therefore, a deep understanding of the roles of frlncRNAs in urologic cancers occurrence and progression will provide novel information for the development of anticancer therapies.
Key words ferroptosis, lncRNA, prostate cancer, proteasome, lysosome
Ferroptosis distinct from apoptosis and refers to iron overload-mediated excessive lipid peroxidation [3]. Interestingly, increasing research on ferroptosis and cancer play critical role in cancer initiation, progression, metastasis and therapeutic resistance. Various cancer-related signaling pathways that regulate ferroptosis demonstrate unique metabolism in cancer cells [4]. Thus, targeting ferroptosis might offer great potential in cancer therapy.
Previous studies have suggested that lncRNAs induce or inhibit ferroptosis of cancer cells in few malignancies including gastric, hepatocellular, and lung cancer [5-7]. Recently, it has been identified that few ferroptosis-related lncRNAs (frlncRNAs) regulate the incidence and progression of urologic malignancies including prostate cancer (PCa), kidney cancer, and bladder cancer (BCa). In this review, we highlight the emerging role of frlncRNAs to predict prognosis of urologic cancers. Moreover, based on current finding on various signaling pathways of the frlncRNAs in urologic cancers we summarize that frlncRNAs regulate ferroptosis-related genes through sponging miRNA, modulating transcription factors and m6A methylation, and alteration in ferroptosis-related proteins through autophagy-lysosomal and proteasome-ubiquitination pathways. We also discuss the roles of metabolism and immune related frlncRNAs in urologic cancers. This review contributes to a comprehensive understanding of the identification and regulation of frlncRNAs in urologic cancers and provide novel information for the development of precise strategies for the treatment of urologic cancers.
Through a ferroptosis-related gene prognostic index (FGPI) analysis combiningg four Gene Expression Omnibus (GEO) database, Feng et al. found that lncRNA PART1 was significantly associated with the prediction biochemical recurrence (BCR) of PCa [8]. Subsequently, ferroptosis-related lncRNA (frlncRNAs) signature was established to predict PCa prognosis, and 5 frlncRNAs (AP006284.1, AC132938.1, BCRP3, AL360181.4 and AL135999.1) were identified for the prediction of PCa BCR [9]. However, validation studies are further required before their progress in clinical application.
Identification of frlncRNAs in kidney cancer
The kidney renal clear cell carcinoma (KIRC) accounts for 80% of cases and the kidney renal papillary cell carcinoma (KIRP) account for 10-15% as two major types of renal cell carcinoma. Through analysis of the RNA-seq count data of KIRC (72 controls vs. 530 cancers) and KIRP (35 controls vs. 291 cancers) and the corresponding clinical information from The Cancer Genome Atlas (TCGA) open database, 5 frlncRNAs were identified as differentially expressed (DOCK8-AS1, SNHG17, RUSC1-AS1, LINC02609, and LUCAT1) and independently correlate with the overall survival (OS) in patients with renal cancer [10]. The differentially expressed frlncRNAs in KIRC have been used to predict the prognosis of KIRC [11, 12]. Based on the ferroptosis-related genes and lncRNAs obtained from the FerrDb and GENCODE databases, a risk assessment model was constructed including 3 frlncRNAs (DUXAP8, LINC02609, and LUCAT1) which significantly correlate with the overall survival of KIRC as an independent factor [13]. This was followed by employing 5 frlncRNAs LINC00460, LINC00894, VPS9D1-AS1, CYTOR, FOXD2-AS1 for construction and validation for the prognostic signature of KIRC [14].
The frlncRNAs also have been identified to predict the prognosis and overall survival of the papillary renal cell carcinoma (PRCC). Through data analysis of PRCC from TCGA, a prognostic signature consisting of 15 frlncRNAs was constructed by Dang et al. [15]. The study found that 7 different frlncRNAs had strong link to the prognosis of patients with PRCC [16]. The frlncRNAs CASC19, AC090197.1, AC099850.3, AL033397.2, LINC00462, and B3GALT1-AS1 were designated as oncogenes in PRCC, whereas LNCTAM34A and AC024022.1 are recognized as tumor suppressor genes in PRCC [17]. These studies have demonstrated that frlncRNAs are associated with the prognosis of kidney cancers, and play diverse roles in cancer progression.
Identification of frlncRNAs in bladder cancer
The frlncRNAs were identified to predict prognosis of bladder cancer (BCa) patients, and a signature composed of 9 frlncRNAs (AL031775.1, AL162586.1, AC034236.2, LINC01004, OCIAD1-AS1, AL136084.3, AP003352.1, Z84484.1, AC022150.2) was constructed [18]. Wang et al. identified 538 differentially expressed frlncRNAs from the TCGA database through co-expression method and differential expression analysis was performed in BCa [19]. In addition, Hou et al. identified 11 differentially expressed frlncRNAs were associated with poor BCa prognosis [20]. However, these two studies did not further investigate the roles of these frlncRNAs in BCa cells. Recently, Liu et al. investigated that AC006160.1 expression was lower in several BCa cell lines BIU-87, T24, RT4, RT-112, and 5637 than in the normal cell line SV-HUC-1, and the overexpression of AC006160.1 significantly inhibited cell proliferation, metastasis, and drug resistance [21]. Through single cell transcriptome sequencing (scRNA-seq), it was indicated that frlncRNAs express specifically in BCa tumor microenvironment, and AL356740.1, LINC02535 and LINC00867 were majorly expressed in tumor cells [22]. These studies showed that frlncRNAs play key roles in the progression of BCa.
Accumulated data suggest that frlncRNAs play critical roles in targeting the regulation of miRNA associated with gene expression. Liu et al. found that the expression of 53 genes potentially interact with frlncRNAs in PCa [9]. Acyl-CoA synthetase long-chain 4 (ACSL4) and solute carrier family 7 membrane 11 (SLC7A11) are common targets that play key roles in ferroptosis. In non-small-cell lung cancer, overexpression of lncRNA NEAT1 can inhibit the expression of ACSL4 but increase the expression of SLC7A11, and decreased ferroptosis and cell apoptosis [23]. In PCa, LncRNA PART1 was speculated to modulate the mRNA expression of endothelial PAS domain containing protein-1 (EPAS1) and ACSL3 through interacting with 60 miRNAs [8]. LncRNA OIP5-AS1 was shown to inhibit ferroptosis of PCa cells after long-term cadmium exposure through miR-128-3p/SLC7A11 signaling [24]. In docetaxel-resistant PCa, lncRNA prostate cancer-associated transcript 1 (PCAT1) was highly expressed, which restrained ferroptosis cell death through c-Myc/miR-25-3p/SLC7A11 signaling [25]. These studies demonstrate that lncRNA promotes progression of PCa by modulating ferroptosis through miRNA and associated genes.
In addition, lncRNAs binding miRNAs also regulate ferroptosis in kidney and bladder cancers. Silencing lncRNA SLC16A1-AS1 induced ferroptosis in ccRCC through miR-143-3p/SLC7A11 signaling [26]. In PRCC, frlncRNAs have been shown to interact with sponge miRNAs and bind proteins to modulate cell proliferation and metastasis [27]. In BCa, LncRNA RP11-89 promotes tumorigenesis and ferroptosis resistance through PROM2-activated iron export by sponging miR-129-5p [28]. The frlncRNAs binding on miRNA to regulate target gene expression results in the progression of urologic cancers. However, additional studies are required.
FrlncRNAs regulates genes through transcription factors and m6A methylation in urologic cancers
As the transcription factors link the lncRNA, target genes, and miRNA, hence the transcription factors might also be involved in frlncRNAs-regulated ferroptosis in cancer cells. Previous studies have shown that the transcription factors such as stem cell factor (SOX2), BTB and CNC homology 1 (BACH1) play important roles in ferroptosis of cancer cells [29, 30]. The signal transduction and activators of transcription 3 (STAT3)-mediated ferroptosis regulate the progression of PCa and kidney cancer [31, 32]. Recently, lncRNAs have been reported to involve in heavy metals induced cancer development. LncRNA lnc-DC mediates arsenic (As)-induced programmed cell death 1 ligand (PD-L1) up-regulation by activating the STAT3 signaling to promote lung cancer [33]. Recent studies suggest that m6A-mediated methylation regulates the expression of oncogenes or tumor suppression genes, which plays important roles in cancer progression. Wang et al. identified 538 differentially expressed frlncRNAs from the TCGA database through co-expression method and differential expression analysis, and found the upstream of 5 lncRNAs might be modified by m6A to modulate their expression in BCa [19]. In addition, Hou et al. identified 11 differentially expressed frlncRNAs were associated with poor BCa prognosis, and m6A methylation-related genes were differentially expressed between the high and low risk groups [20]. However, there is still a large gap in understanding as how frlncRNAs regulate ferroptosis related genes, sponging miRNA and the transcription factors, as well as m6A methylation in urologic cancer.
Regulation of frlncRNAs in urologic cancers by protein levels
Studies show that lncRNAs can regulate target effectors at the protein levels. The autophagy-lysosomal pathway and proteasome-mediated ubiquitination-mediated degradation are two major processes in regulating protein levels. Our previous studies have showed that lncRNA FAM66C can regulate PCa cells proliferation and metastasis through autophagy-lysosomal pathway and proteasome-ubiquitination pathways [34, 35]. In human pancreatic ductal adenocarcinoma (PDAC) cells, STAT3 mediated lysosomal cysteine protease cathepsin B expression to induce ferroptosis [36]. Therefore, ferroptosis is also proposed as a process of autophagy-lysosomal cell death. LncRNA HEPFAL was found to increase the sensitivity of erastin-induced ferroptosis in hepatocellular carcinoma, and induce ubiquitination of SLC7A11 and decrease its expression by reducing the stability of SLC7A11 protein [7]. The voltage-dependent anion channel (VDAC)-mediated mitochondria dysfunction is necessary for erastin-induced ferroptosis and VDAC3 can act as a directly target of erastin [37]. The lncRNA BDNF-AS/WDR5/FBXW7 axis mediates ferroptosis in gastric cancer peritoneal metastasis by regulating VDAC3 ubiquitination [38]. In colorectal cancer, lncRNA LINC00239 inhibits ferroptosis by binding to Keap1 to stabilize the transcription factor Nrf2 [39]. The lncRNA can also be stabilized by RNA binding protein. It is reported that high-density lipoprotein-binding protein (HDLBP)-stabilized lncRNA lncFAL reduces ferroptosis by inhibiting degradation of ferroptosis suppressor protein 1 (FSP1) in hepatocellular carcinoma [40]. It also showed that lncRNAs drive chemoresistance and metastasis by regulating ubiquitination and promote PCa cell growth [35, 41, 42]. All these studies suggested that regulations of frlncRNAs on ferroptosis-related genes and proteins may control the development of urologic cancers, but the underlying mechanism needs to be further elucidated (Table 1).
|
Table 1. Summary of regulation of ferroptosis-relative lncRNAs in urologic cancers. |
|||||
|
Urologic Cancers |
LncRNA |
Pathway |
Target |
Associated Effects |
Reference |
|
Pca |
PART1 |
60miRNAs |
mRNA:EPAS1, ACSL3 |
Inhibited ferroptosis, Increased biochemical recurrence and radiation resistance |
[8] |
|
Pca |
OIP5-AS1 |
miR-128-3p |
mRNA:SLC7A11 |
Ferroptosis resistance, Promoted cell growth |
[24] |
|
Pca |
PCAT1 |
c-Myc/miR-25-3p |
mRNA:SLC7A11 |
Restrained ferroptosis, Increased docetaxel-resistant |
[25] |
|
ccRCC |
SLC16A1-AS1 |
miR-143-3p |
mRNA:SLC7A11 |
Induced ferroptosis, Inhibited cell viability, proliferation, and migration |
[26] |
|
Bca |
RP11-89 |
sponging miR-129-5p |
mRNA:PROM2 |
Ferroptosis resistance, Promotes tumorigenesis |
[28] |
|
PCa Kidney cancer |
Unknown |
Unknown |
STAT3 |
Mediated ferroptosis, Regulated the progression of PCa and kidney cancer |
[31, 32] |
|
BCa |
AC096921.2, LINC02762, etc. |
m6A methylation |
m6A methylation related gene |
Induced or inhibited ferroptosis, Protective or poor BCa prognosis |
[19] |
|
BCa |
AL583785.1, LINC02762, etc. |
m6A methylation |
m6A methylation related gene |
Different between the high and low risk groups, predicting the prognosis of BCa |
[20] |
|
PCa |
HOTAIR |
E3 ubiquitin |
MDM2 |
Castration-resistant |
[41] |
|
PCa |
PCBP1-AS1 |
AR/AR-V7 deubiquitination |
USP22-AR/AR-V7 |
Castration-resistant |
[42] |
|
Pca |
NEAT1 |
Sponging miR-34a-5p and miR-204-5p |
mRNA:ACSL4 |
Increased oxidation of fatty acids, docetaxel resistance |
[46] |
|
PCa |
AP006284.1, AC132938.1, etc. |
Unknown |
Unknown |
Immune cell infiltration |
[8, 9] |
|
ccRCC |
AC026401.3, LINC01615, etc. |
CD80, IDO1, and LAG3 |
Unknown |
Immune cell infiltration and changed immune microenvironment |
[12, 50-53] |
|
PRCC |
ZFAS1, AC010624.2, etc. |
Unknown |
CD80, IDO1, and LAG3 |
Immune cell infiltration |
[15] |
|
PRCC |
CASC19, AC090197.1, etc. |
Unknown |
CD160, TNFSF4, CD80, BTLA, and TNFRSF9 |
Predicted the overall survival outcome |
[17] |
|
Bca |
High risk group lncRNAs |
IL-17 signaling pathway and TNF signaling pathway, |
PDCD-1 (PD-1), CTLA4, and LAG3 |
Predicted immune and tumor-related pathways |
[20, 56] |
Based on TCGA database, it is revealed that 9 frlncRNAs signature that mediate T cell functions, such as cytolytic activity, human leukocyte antigen activity, inflammation regulation, and type II interferon response coordination are significantly different between high- and low- risk levels of ccRCC and may affect its prognosis [12]. Dong et al. analyzed the frlncRNAs signature with ccRCC prognostication, and showed that the immune checkpoint CD44, TNFRSF18, TNFSF14, TNFRSF8, CD276, and TNFRSF25 were upregulated in the high-risk group, while HAVCR2, NRP1, and HHLA2 were upregulated in the low-risk group [54]. In PRCC, 15 frlncRNAs were identified to predict the prognostic signature. Moreover, the frlncRNAs constructed high-risk group had a greater degree of immune cell infiltration than the low-risk group. A significantly higher expression level of immune checkpoints including CD80, IDO1, and LAG3 was shown in the high-risk group than in the low-risk group [15]. Wu et al. found some frlncRNAs targeting inflammation- and immune- promoting genes viz. CD160, TNFSF4, CD80, BTLA, and TNFRSF9 differentially expressed in the high- and low- risk groups with PRCC [17]. It indicates that similar and dissimilar frlncRNAs-related immune genes were found in different studies, and the accurate molecular functions still needs to further verification.
In addition, frlncRNAs also predict immune infiltration and immunotherapeutic outcomes in BCa [55]. The frlncRNAs predicted immune- and tumor- related pathways such as IL-17 signaling and TNF signaling, and the immune checkpoints such as PDCD-1 (PD-1), CTLA4, and LAG3, were differentially expressed between the high- and low- BCa risk groups [20, 56]. The high-risk BCa group was positively associated with tumor-infiltrating immune cells, including monocytes, fibroblasts, and macrophages, but negatively related to CD4+ T cells and CD8+ T cells [57]. Based on the frlncRNAs related immune landscape, developing anti-BCa mRNA vaccine facilities the individual precision treatment of BCa patients [58].
To summarize, the mechanistic relationships between ferroptosis and lncRNAs regulation in cancer progression is a recent topic of interest in cancer research. The frlncRNAs are involved in urologic cancers progression through various pathways including metabolic pathways (lipid peroxidation), and tumor microenvironment, and immune cell infiltration. Interestingly, frlncRNAs does not only regulate ferroptosis-related signaling and genes by sponging miRNA and regulating transcription factors and m6A methylation, but also control protein levels through autophagy-lysosomal and proteasome-ubiquitination degradation pathways (Figure 1). Certainly, there still a lot unknowns in the frlncRNAs network. Therefore, deeper understanding of how the frlncRNAs affect urologic cancer cells through integrated analysis of the network of frlncRNAs with miRNA, mRNA, targeted genes, transcription factors, metabolic pathways, and autophagy-lysosomal and proteasome-ubiquitination pathways, which will help to obtain novel discovery of anticancer therapies.
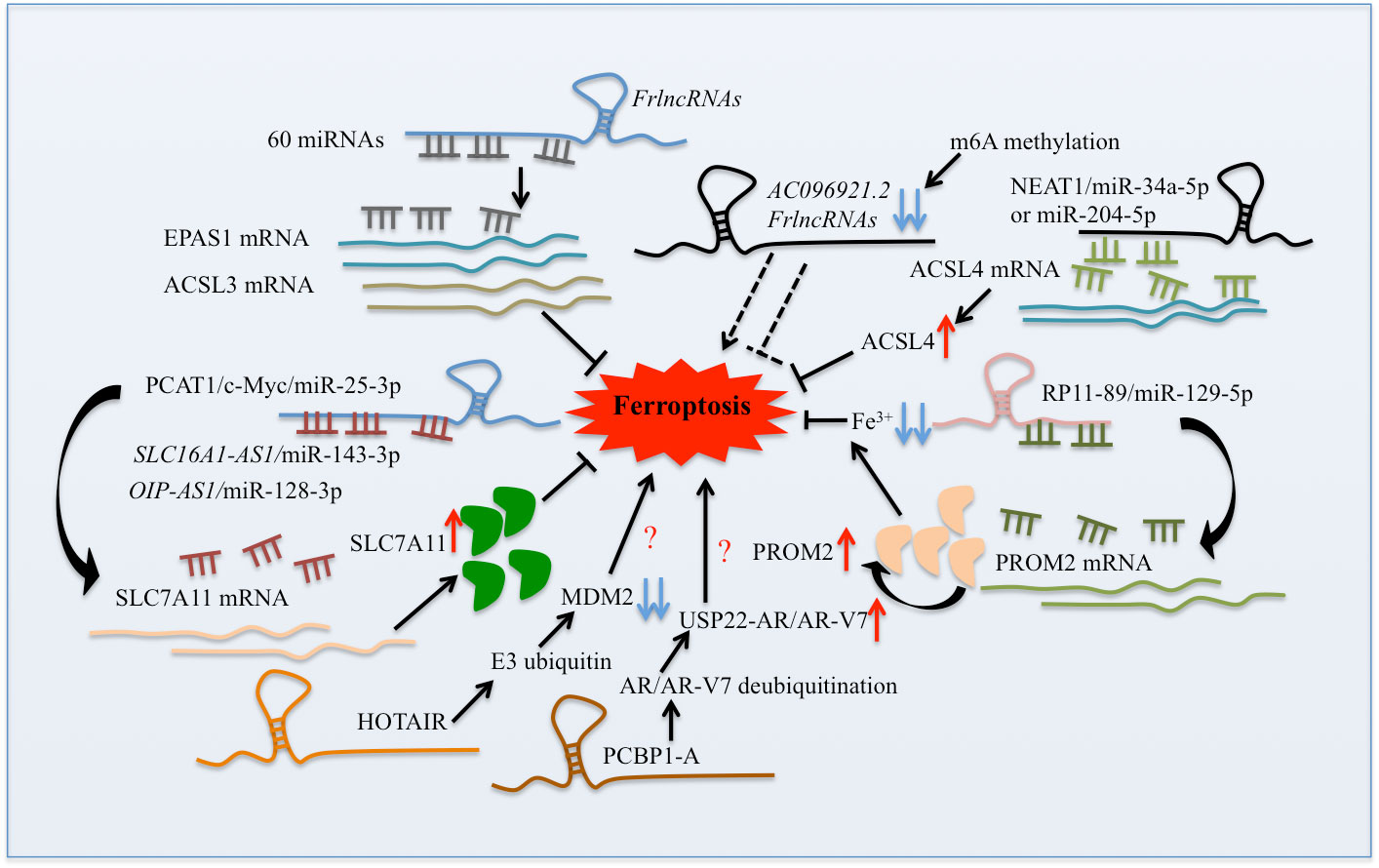 Figure 1. The mechanism of lncRNAs regulated ferroptosis urologic cancers. The lncRNAs could not only regulate ferroptosis-related genes and proteins by sponging miRNA, activating m6A methylation, and regualating ubiquitination pathway ( Modified from Qu et al., 2022 [2] ).
Figure 1. The mechanism of lncRNAs regulated ferroptosis urologic cancers. The lncRNAs could not only regulate ferroptosis-related genes and proteins by sponging miRNA, activating m6A methylation, and regualating ubiquitination pathway ( Modified from Qu et al., 2022 [2] ).
This work was supported by the Foundation of Health and Family Planning Commission of Wuxi City (Q201807), the Foundation of Clinical Medical Science and Technology Development project of Jiangsu University (JLY20180005), the Foundation of Science and Technology Development Medical and Health Guidance Plan Project of Wuxi City (2020061), the Innovation Foundation of Science and Technology Bureau of Yixing City (2020SF08), the Municipal Health Commission Suitable Technology promotion project of Wuxi City (T202126), and the Municipal Health Commission Suitable Technology promotion project of Jiangsu province (LSD2022008).
Ethical policy
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study. Approval from institutional ethical committee was taken.
Availability of data and materials
All data generated or analysed during this study are included in this publication.
Author contributions
WX, JG, ZQ and YX discussed the topic of this article. WX and JG wrote the draft of the manuscript. WX participated in interpretation of the literature. JG, ZQ and YX revised the review. All authors revised the draft of the manuscript. All authors have read and agreed to the published version of the manuscript.
Competing interests
All authors declare no competing interests.
Funding
None.
- Huang J, Wang J, He H, Huang Z, Wu S, Chen C, Liu W, Xie L, Tao Y, Cong L et al: Close interactions between lncRNAs, lipid metabolism and ferroptosis in cancer. Int J Biol Sci 2021, 17(15): 4493-4513.
- Qu L, He X, Tang Q, Fan X, Liu J, Lin A: Iron metabolism, ferroptosis, and lncRNA in cancer: knowns and unknowns. J Zhejiang Univ Sci B 2022, 23(10): 844-862.
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS et al: Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012, 149(5): 1060-1072.
- Lei G, Zhuang L, Gan B: Targeting ferroptosis as a vulnerability in cancer. Nat Rev Cancer 2022, 22(7): 381-396.
- Lin Z, Song J, Gao Y, Huang S, Dou R, Zhong P, Huang G, Han L, Zheng J, Zhang X et al: Hypoxia-induced HIF-1alpha/lncRNA-PMAN inhibits ferroptosis by promoting the cytoplasmic translocation of ELAVL1 in peritoneal dissemination from gastric cancer. Redox Biol 2022, 52: 102312.
- Zhang R, Pan T, Xiang Y, Zhang M, Xie H, Liang Z, Chen B, Xu C, Wang J, Huang X et al: Curcumenol triggered ferroptosis in lung cancer cells via lncRNA H19/miR-19b-3p/FTH1 axis. Bioact Mater 2022, 13: 23-36.
- Meng X, Xiao W, Sun J, Li W, Yuan H, Yu T, Zhang X, Dong W: CircPTK2/PABPC1/SETDB1 axis promotes EMT-mediated tumor metastasis and gemcitabine resistance in bladder cancer. Cancer Lett 2022, 554: 216023.
- Feng D, Shi X, Xiong Q, Zhang F, Li D, Wei W, Yang L: A Ferroptosis-Related Gene Prognostic Index Associated With Biochemical Recurrence and Radiation Resistance for Patients With Prostate Cancer Undergoing Radical Radiotherapy. Front Cell Dev Biol 2022, 10: 803766.
- Liu C, Gao Y, Ni J, Chen S, Hu Q, Wang C, Hu M, Chen M: The ferroptosis-related long non-coding RNAs signature predicts biochemical recurrence and immune cell infiltration in prostate cancer. BMC Cancer 2022, 22(1): 788.
- Shu X, Zhang Z, Yao ZY, Xing XL: Identification of Five Ferroptosis-Related LncRNAs as Novel Prognosis and Diagnosis Signatures for Renal Cancer. Front Mol Biosci 2021, 8: 763697.
- Liu JW, Supandi F, Dhillon SK: Ferroptosis-Related Long Noncoding RNA Signature Predicts Prognosis of Clear Cell Renal Carcinoma. Folia Biol (Praha) 2022, 68(1): 1-15.
- Bai Z, Zhao Y, Yang X, Wang L, Yin X, Chen Y, Lu J: A Novel Prognostic Ferroptosis-Related Long Noncoding RNA Signature in Clear Cell Renal Cell Carcinoma. J Oncol 2022, 2022: 6304824.
- Xing XL, Yao ZY, Ou J, Xing C, Li F: Development and validation of ferroptosis-related lncRNAs prognosis signatures in kidney renal clear cell carcinoma. Cancer Cell Int 2021, 21(1): 591.
- Zhu Z, Zhang C, Qian J, Feng N, Zhu W, Wang Y, Gong Y, Li X, Lin J, Zhou L: Construction and validation of a ferroptosis-related long noncoding RNA signature in clear cell renal cell carcinoma. Cancer Cell Int 2022, 22(1): 283.
- Dang R, Jin M, Nan J, Jiang X, He Z, Su F, Li D: A Novel Ferroptosis-Related lncRNA Signature for Prognosis Prediction in Patients with Papillary Renal Cell Carcinoma. Int J Gen Med 2022, 15: 207-222.
- Tang X, Jiang F, Wang X, Xia Y, Mao Y, Chen Y: Identification of the Ferroptosis-Related Long Non-Coding RNAs Signature to Improve the Prognosis Prediction in Papillary Renal Cell Carcinoma. Front Surg 2022, 9: 741726.
- Wu Z, Huang X, Cai M, Huang P: Potential biomarkers for predicting the overall survival outcome of kidney renal papillary cell carcinoma: an analysis of ferroptosis-related LNCRNAs. BMC Urol 2022, 22(1): 152.
- Chen M, Nie Z, Li Y, Gao Y, Wen X, Cao H, Zhang S: A New Ferroptosis-Related lncRNA Signature Predicts the Prognosis of Bladder Cancer Patients. Front Cell Dev Biol 2021, 9: 699804.
- Wang Y, Zhang S, Bai Y, Li G, Wang S, Chen J, Liu X, Yin H: Development and Validation of Ferroptosis-Related LncRNA Biomarker in Bladder Carcinoma. Front Cell Dev Biol 2022, 10: 809747.
- Hou J, Lu Z, Cheng X, Dong R, Jiang Y, Wu G, Qu G, Xu Y: Ferroptosis-related long non-coding RNA signature predicts the prognosis of bladder cancer. BMC Cancer 2022, 22(1): 719.
- Liu J, Cui J, Zhao S, Wu M, Wang J, Zhang Y, Jin B, Wang J: Ferroptosis-Related Long Noncoding RNAs Have Excellent Predictive Ability for Multiomic Characteristics of Bladder Cancer. Oxid Med Cell Longev 2022, 2022: 9316847.
- Xiang X, Guo Y, Chen Z, Zhang F, Huang J, Qin Y: A prognostic risk prediction model based on ferroptosis-related long non-coding RNAs in bladder cancer: A bulk RNA-seq research and scRNA-seq validation. Medicine (Baltimore) 2022, 101(51): e32558.
- Wu H, Liu A: Long non-coding RNA NEAT1 regulates ferroptosis sensitivity in non-small-cell lung cancer. J Int Med Res 2021, 49(3): 300060521996183.
- Zhang Y, Guo S, Wang S, Li X, Hou D, Li H, Wang L, Xu Y, Ma B, Wang H et al: LncRNA OIP5-AS1 inhibits ferroptosis in prostate cancer with long-term cadmium exposure through miR-128-3p/SLC7A11 signaling. Ecotoxicol Environ Saf 2021, 220: 112376.
- Fang P, Jiang Q, Liu S, Gu J, Hu K, Wang Z: Circ_0002099 is a novel molecular therapeutic target for bladder cancer. Drug Dev Res 2022, 83(8): 1890-1905.
- Li YZ, Zhu HC, Du Y, Zhao HC, Wang L: Silencing lncRNA SLC16A1-AS1 Induced Ferroptosis in Renal Cell Carcinoma Through miR-143-3p/SLC7A11 Signaling. Technol Cancer Res Treat 2022, 21: 15330338221077803.
- Trevisani F, Floris M, Vago R, Minnei R, Cinque A: Long Non-Coding RNAs as Novel Biomarkers in the Clinical Management of Papillary Renal Cell Carcinoma Patients: A Promise or a Pledge? Cells 2022, 11(10).
- Luo W, Wang J, Xu W, Ma C, Wan F, Huang Y, Yao M, Zhang H, Qu Y, Ye D et al: LncRNA RP11-89 facilitates tumorigenesis and ferroptosis resistance through PROM2-activated iron export by sponging miR-129-5p in bladder cancer. Cell Death Dis 2021, 12(11): 1043.
- Wang X, Chen Y, Wang X, Tian H, Wang Y, Jin J, Shan Z, Liu Y, Cai Z, Tong X et al: Stem Cell Factor SOX2 Confers Ferroptosis Resistance in Lung Cancer via Upregulation of SLC7A11. Cancer Res 2021, 81(20): 5217-5229.
- Stockwell BR, Jiang X: A Physiological Function for Ferroptosis in Tumor Suppression by the Immune System. Cell Metab 2019, 30(1): 14-15.
- Ji J, Li H, Wang W, Yuan B, Shen T: ARPC1A is regulated by STAT3 to inhibit ferroptosis and promote prostate cancer progression. Hum Cell 2022, 35(5): 1591-1601.
- Li Y, Zhang Y, Qiu Q, Wang L, Mao H, Hu J, Chen Z, Du Y, Liu X: Energy-Stress-Mediated AMPK Activation Promotes GPX4-Dependent Ferroptosis through the JAK2/STAT3/P53 Axis in Renal Cancer. Oxid Med Cell Longev 2022, 2022: 2353115.
- Shen C, Li Z, Zhang Y, Zhang Z, Wu Z, Da L, Yang S, Wang Z, Zhang Y, Qie Y et al: Identification of a dysregulated CircRNA-associated gene signature for predicting prognosis, immune landscape, and drug candidates in bladder cancer. Front Oncol 2022, 12: 1018285.
- Yuan ZQKLJGYXX: Long Non-coding RNA FAM66C Promotes Prostate Cancer Metastasis via JNK Mediated Proteasome and Lysosomal Pathway. Annals of Urologic Oncology 2022 Epub ahead of print.
- Xie Y, Gu J, Qin Z, Ren Z, Wang Y, Shi H, Chen B: Long non-coding RNA FAM66C is associated with clinical progression and promotes cell proliferation by inhibiting proteasome pathway in prostate cancer. Cell Biochem Funct 2020, 38(8): 1006-1016.
- Gao H, Bai Y, Jia Y, Zhao Y, Kang R, Tang D, Dai E: Ferroptosis is a lysosomal cell death process. Biochem Biophys Res Commun 2018, 503(3): 1550-1556.
- Wu Y, Zhang S, Gong X, Tam S, Xiao D, Liu S, Tao Y: The epigenetic regulators and metabolic changes in ferroptosis-associated cancer progression. Mol Cancer 2020, 19(1): 39.
- Huang G, Xiang Z, Wu H, He Q, Dou R, Lin Z, Yang C, Huang S, Song J, Di Z et al: The lncRNA BDNF-AS/WDR5/FBXW7 axis mediates ferroptosis in gastric cancer peritoneal metastasis by regulating VDAC3 ubiquitination. Int J Biol Sci 2022, 18(4): 1415-1433.
- Han Y, Gao X, Wu N, Jin Y, Zhou H, Wang W, Liu H, Chu Y, Cao J, Jiang M et al: Long noncoding RNA LINC00239 inhibits ferroptosis in colorectal cancer by binding to Keap1 to stabilize Nrf2. Cell Death Dis 2022, 13(8): 742.
- Yuan J, Lv T, Yang J, Wu Z, Yan L, Yang J, Shi Y: HDLBP-stabilized lncFAL inhibits ferroptosis vulnerability by diminishing Trim69-dependent FSP1 degradation in hepatocellular carcinoma. Redox Biol 2022, 58: 102546.
- Zhang A, Zhao JC, Kim J, Fong KW, Yang YA, Chakravarti D, Mo YY, Yu J: LncRNA HOTAIR Enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell Rep 2015, 13(1): 209-221.
- Zhang B, Zhang M, Shen C, Liu G, Zhang F, Hou J, Yao W: LncRNA PCBP1-AS1-mediated AR/AR-V7 deubiquitination enhances prostate cancer enzalutamide resistance. Cell Death Dis 2021, 12(10): 856.
- Doll S, Proneth B, Tyurina YY, Panzilius E, Kobayashi S, Ingold I, Irmler M, Beckers J, Aichler M, Walch A et al: ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol 2017, 13(1): 91-98.
- Alghandour R, Ebrahim MA, Elshal AM, Ghobrial F, Elzaafarany M, MA EL: Repurposing metformin as anticancer drug: Randomized controlled trial in advanced prostate cancer (MANSMED). Urol Oncol 2021, 39(12): 831 e831-831 e810.
- Schonenberger MJ, Krek W, Kovacs WJ: EPAS1/HIF-2alpha is a driver of mammalian pexophagy. Autophagy 2015, 11(6):967-969.
- Jiang X, Guo S, Zhang Y, Zhao Y, Li X, Jia Y, Xu Y, Ma B: LncRNA NEAT1 promotes docetaxel resistance in prostate cancer by regulating ACSL4 via sponging miR-34a-5p and miR-204-5p. Cell Signal 2020, 65: 109422.
- Wang W, Green M, Choi JE, Gijon M, Kennedy PD, Johnson JK, Liao P, Lang X, Kryczek I, Sell A et al: CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 2019, 569(7755): 270-274.
- Shi L, Liu Y, Li M, Luo Z: Emerging roles of ferroptosis in the tumor immune landscape: from danger signals to anti-tumor immunity. FEBS J 2022, 289(13): 3655-3665.
- Han Z, Wang H, Liu Y, Xing XL: Establishment of a prognostic ferroptosis- and immune-related long noncoding RNAs profile in kidney renal clear cell carcinoma. Front Genet 2022, 13: 915372.
- Chen X, Tu J, Ma L, Huang Y, Yang C, Yuan X: Analysis of Ferroptosis-Related LncRNAs Signatures Associated with Tumor Immune Infiltration and Experimental Validation in Clear Cell Renal Cell Carcinoma. Int J Gen Med 2022, 15: 3215-3235.
- Zhou Z, Yang Z, Cui Y, Lu S, Huang Y, Che X, Yang L, Zhang Y: Identification and Validation of a Ferroptosis-Related Long Non-Coding RNA (FRlncRNA) Signature to Predict Survival Outcomes and the Immune Microenvironment in Patients With Clear Cell Renal Cell Carcinoma. Front Genet 2022, 13: 787884.
- Ju L, Shi Y, Liu G: Identification and validation of a ferroptosis-related lncRNA signature to robustly predict the prognosis, immune microenvironment, and immunotherapy efficiency in patients with clear cell renal cell carcinoma. PeerJ 2022, 10: e14506.
- Wei SY, Feng B, Bi M, Guo HY, Ning SW, Cui R: Construction of a ferroptosis-related signature based on seven lncRNAs for prognosis and immune landscape in clear cell renal cell carcinoma. BMC Med Genomics 2022, 15(1): 263.
- Dong Y, Liu D, Zhou H, Gao Y, Nueraihemaiti Y, Xu Y: A Prognostic Signature for Clear Cell Renal Cell Carcinoma Based on Ferroptosis-Related lncRNAs and Immune Checkpoints. Front Genet 2022, 13: 912190.
- Zhou R, Liang J, Tian H, Chen Q, Yang C, Liu C: Development of a Ferroptosis-Related lncRNA Signature to Predict the Prognosis and Immune Landscape of Bladder Cancer. Dis Markers 2021, 2021: 1031906.
- Liu J, Zhang Z, Liu X, Zhang W, Meng L, Wang J, Lv Z, Xia H, Zhang Y, Wang J: Predictive role of ferroptosis-related long non-coding RNAs in bladder cancer and their association with immune microenvironment and immunotherapy response. World J Surg Oncol 2022, 20(1): 47.
- Li X, Zhou L, Lu T, Zhang L, Li Y, Xu J, Yin M, Long H: Constructing an immune- and ferroptosis-related lncRNA signature to predict the immune landscape of human bladder cancer. J Clin Lab Anal 2022, 36(5): e24389.
- Gui CP, Li JY, Fu LM, Luo CG, Zhang C, Tang YM, Zhang LZ, Shu GN, Wu RP, Luo JH: Identification of mRNA vaccines and conserved ferroptosis related immune landscape for individual precision treatment in bladder cancer. J Big Data 2022, 9(1): 88.
Annals of urologic oncology
p-ISSN: 2617-7765, e-ISSN: 2617-7773
 Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript