11 Nov 2023
Issue 4
Views:2557
Downloads:19
Review Article | Open Access
Research Progress on the Mechanism of Androgen Receptor Signaling Pathway in Castration-Resistant Prostate Cancer
Na Cui1, Mingjie Li1
13201 Hospital, Hanzhong City, Shaanxi Province, 723000, P.R. China.
Correspondence: Mingjie Li (3201 Hospital, Tianhan Avenue 783, Hanzhong City, Shaanxi Province, 723000, P.R. China; Email: 1273931857@qq.com).
Annals of Urologic Oncology 2023, 6(4): 132-138. https://doi.org/10.32948/auo.2023.11.09
Received: 14 Oct 2023 | Accepted: 30 Oct 2023 | Published online: 11 Nov 2023
Abstract
Prostate cancer (Pca) remains the most common malignancy worldwide in men, and the second leading cause of mortality only to lung cancer. Besides surgery, androgen deprivation therapy (ADT) is a major treatment for Pca. However, ADT leads to the inevitable progression of castration-resistant Pca (CRPC). The transition from hormone-dependent Pca (ADPC) to CRPC has been shown to involve reactivation of the androgen receptor (AR) signaling pathway. The evidence become strong that Pca develop adaptive mechanisms for maintaining AR signaling to allow for survival and further evolution. This article mainly reviews the research progress of the mechanism(s) of AR signaling in CRPC and provides scientific basis and new ideas for the diagnosis and treatment of this phenotype.
Key words androgen receptor, signaling pathway, castration-resistant prostate cancer, mechanism of action
Introduction
At present, prostate cancer (Pca) is the most common malignancy of the males in the world. According to the estimate by the American Cancer Society, its incidence ranks first place in men, and its mortality rate is only second to lung cancer. According to relevant surveys, the incidence and mortality of Pca are increasing [1-3]. Androgen deprivation therapy (ADT) is a major treatment modality for advance-stage Pca where serum testosterone is depleted by pharmaceutical or surgical castration [4]. Although ADT can significantly inhibit the growth and proliferation of tumors in the early stage, the growth of tumors will eventually be uncontrolled. ADT gradually becomes ineffective, and the disease develops into castrate-resistant Pca (CRPC) [5, 6]. The growth of Pca is stimulated by, the androgen receptor (AR) that binds to ligands such as dihydrotestosterone (DHT) modulates the expression of target genes [7, 8]. A large number of studies have found that AR signaling pathway plays an important role in promoting the transformation of androgen dependent Pca (ADPC) to CRPC [9]. The reactivation of AR signaling pathway is closely related to this transition, in addition, there are other AR-related changes that promote the occurrence of CRPC [10]. This article mainly reviews the research progress of the mechanism ofAR signaling pathway in CRPC and provides scientific basis and new ideas for its diagnosis and treatment.
The basic structure of AR gene and AR signaling pathway
The AR gene located on Xq11-13 is composed of 8 exons, which encode the N-terminal domain (NTD), DNA binding domain (DBD), hinge domain, ligand binding domain (LBD) [11]. AR is a member of the nuclear receptor superfamily, which acts as a transcription factor [12]. In the absence of hormones, AR binds to heat shock proteins and is located in the cytoplasm in a non-active conformation. After androgen binding, AR rapidly undergoes conformational changes and shuttles to the nucleus and binds to the androgen response element (ARE) [13]. Recruitment of coactivators and corepressors, as well as chromatin remodeling, promote the induction of AR dependent gene transcription [14]. Specific combinations of co-factors recruited to ARE provide tissue and ligand-specific factors such as prostate specific antigen (PSA) and transmembrane protease expression [15].
AR genetic mutation
AR genetic mutations have been found to be rare in Pca at an early stage or without hormone therapy, whereas AR genetic mutations are common in CRPC. The mechanism of abrupt change of AR leading to CRPC can be summarized as AR conformational change, which not only depends on androgens, but also can combine with other similar hormones. The conformation of AR can be disrupted by point mutations, which usually occur in CRPC and mediate resistance to AR targeted therapy [16, 17]. Abrupt changes in LBD allow AR to be activated by antiandrogens or other steroid hormones [18]. For example, LNCaP cells with AR mutation T877A can be activated by flutamide, estrogen, and progesterone [19]. AR with L701H/T877A double abrupt change can be activated by glucocorticoids [20]. Studies have shown that Pca cells with AR mutations are better able to adapt to ADT and survive, so that cancer cells harboring AR aberrations can play a role without relying on androgens, but can be activated with estrogen, glucocorticoids and even anti-androgen drugs, thus promoting hormone resistance and cancer progression [21, 22]. This androgen-independent AR signaling pathway is also known as the ligand-independent pathway.
AR gene and its enhancer amplification
Amplification of AR loci is one of the most common mechanisms of castration resistance, occurring in 50% of men with CRPC, and they typically contain enhancers located about 700 kilobases upstream [23, 24]. In some tumors, AR genes and enhancers expand independently of each other [25]. AR enhancers bind to transcriptional activators including FOXA1, GATA2, NKX3.1, HOXB13, and AR itself. Amplification of AR and its enhancers was associated with higher levels of AR expression [26]. Zhou et al. [27] found that highly repetitive amplicons contain intergenic regulatory elements that interact with AR genes in CRPC, and AR enhancers are critical for cell viability in metastatic LNCaP Pca cell lines. In addition, the study found that unamplified AR was detected in only 2 out of 205 untreated Pca [28]. In cases of metastatic CRPC, high levels of AR gene amplification were found in 38% to 63% of circulating tumor cells [29], suggesting that selective AR amplification occurs during CRPC progression. Overexpression caused by AR gene and enhancer amplification promotes the conversion of ADPC to CRPC and the progression of CRPC.
AR splicing variants
Seventeen AR variants that lack LBD have been discovered in the last 10 years [30]. The formation mechanism can be summarized as the high expression of various splicing factors in CRPC [31-34]. The enhanced expression of splicing factors will promote their recruitment to pre-mRNA, thus facilitating the mRNA splicing process. Therefore, altering the splicing process leads to dysregulation of splicing process. Splicing factors, which are rich in proline and glutamine, are responsible for widespread upregulation of spliceosomal gene expression in CRPC, thus activating multiple carcinogenic pathways including AR. Since splicing variants lack LBD, they remain unbound by most AR signaling inhibitors and do not depend on androgen binding, but directly activate the expression of AR driver genes (
Figure 1). We compared the morphological differences of 19 patients with primary hormone sensitivity and CRPC, and evaluated the expression of AR-FL, AR-V7, AR-V4, ARv567es, AR-V3 and AR8 mRNAs. AR-FL, AR-V7, ARv567es and AR-V3 mRNAs were expressed in hormone-sensitive prostate cancer (HSPC), AR-V3 mRNA expression was significantly increased in CRPC, accounting for 81.2% (13/16) [35]. AR-FL positively correlated with AR-V7 (r= 0.93, P< 0.001), ARv567es (r= 0.72, P< 0.001) and AR-V3 (r= 0.81, P< 0.001) mRNA expression, which confirmed that AR-V7 was expressed at a low level in HSPC. The expression of AR-V7 increased in CRPC. The expression of AR-V7 protein in CRPC, human nucleus pulposus cell and benign tumor samples was evaluated by immunohistochemistry. The results showed that nucleolar AR-V7 staining was positive in 44% of CRPC samples, compared with 9% in human nucleus pulposus cell and 0 in benign tumor samples. This study further evaluated the predictive value of AR-V7, finding that high levels of AR-V7 cytoplasmic staining were associated with a greater risk of PSA recurrence after radical prostatectomy [36]. These results suggest that AR splicing variants are significantly higher in CRPC than human nucleus pulposus cells, and their high expression is closely related to castration-resistant formation and progression of CRPC.
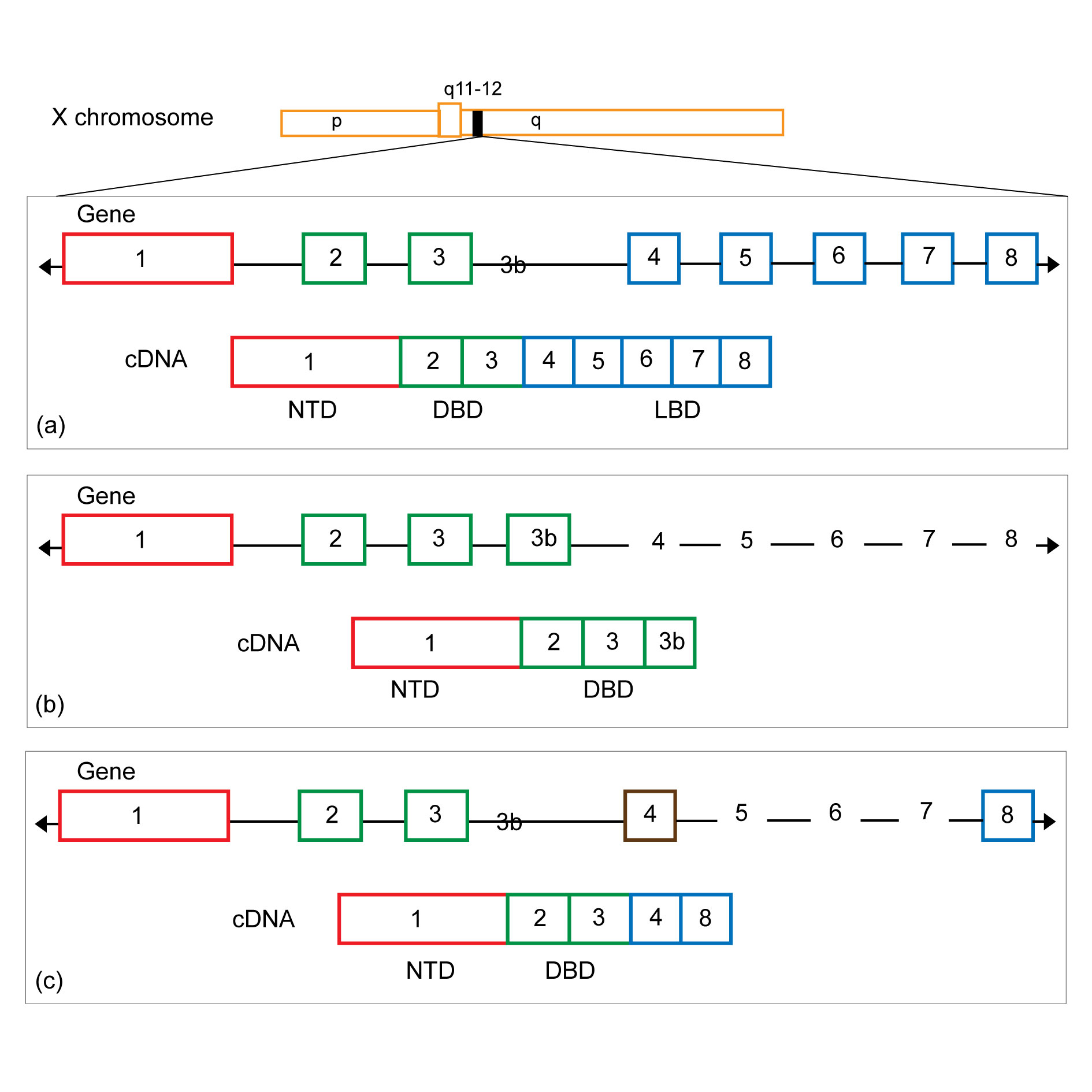 Figure 1.
Figure 1. Schematic structure of human androgen receptor (AR) and AR splice variant 7 (AR-V7) and 567(AR567es). (a) The full-length AR; (b) AR-V7 (also named AR3) encodes a protein with exons 1-3 and a terminal cryptic exon (CE3); (c) AR567es encodes a protein comprised of exons 1-4, and because of a frame-shift due to loss of exons 5-7, exon 8 has a stop codon generated after the first 10 amino acids resulting in a shortened exon 8.
AR co-regulatory factor
AR translocations to the cell nucleus with androgen binding and ARE binding, recruitment of coregulated node factors and chromatin remodeling promote the induction of AR dependent gene transcription, and recruitment of specific combinations of ARE coregulated node factors can provide tissue and ligand-specific gene expression. Recruitment of coregulatory factor is a critical regulatory step in AR signaling [37], which alters the transcription process by enhancing or decreasing the transcriptional activity of AR. For example, in the presence of weaker androgen androstenedione, the increased expression of steroid receptor coactivator 1 (SRC1) and TIF2 can stimulate AR activity. The expression of MAGEA-11 coactivator recruited through the AR NH2- terminal FxxLF motif is increased in CRPC [38]. Primate-specific MAGE-A11 was found to be one of the coactivator, chromatin immunoprecipitation revealed that AR recruits intron 10 of the FSTL1 gene, which contains the classic ARE, and AR co-upregulates FSTL1 with MAGE-A11 to promote the growth and progression of CRPC [39]. Related drugs that block coactivator recruitment are also being developed and are attractive targets because they are mediated by protein-protein interactions, and this antagonist can overcome resistance to conventional antiandrogens and remain effective in advanced Pca. Several binding domains outside ABS have been explored to block AR-coactivator interactions and inhibit AR signaling. Within the AR domain, the most promising compounds have been produced at the AF-1, AF-2 and BH-3 sites [40].
Long non-coding RNAs
Studies have shown that only a very small number of total genome sequences in humans encode protein-coding transcripts, while the rest are actively transcribed to generate large amounts of functional non-coding RNAs [41-44]. Long non-coding RNAs (lncRNAs) are defined as those with lengths greater than 200 nucleotides. Abhijit Parolia et al. demonstrated that HORAS5 is a stable, cytoplasmic lncRNA that promotes CRPC proliferation and survival by maintaining AR activity under androgen-depleted conditions [45]. The most typical role of lncRNA is to act as epigenetic modulator in gene regulation and promote the progression of CRPC by acting on AR signaling pathway. The overexpression of nuclear-enriched abundant transcript1 (NEAT1), which regulates gene expression through epigenetic regulation, has been associated with aggressive Pca. Lin [46] found that NEAT1 is up-regulated by estrogen receptor α (ERα), and overexpression of NEAT1 changes the levels of H3K4me3 and H3AcK9 by directly binding to histone H3, thus altering the chromatin distribution of the target promoter, thereby promoting active transcription. Given that estrogen signaling via ERα bypasses the androgen signaling pathway, ERα-Neat1-mediated chromatin alterations may drive tumor growth and CRPC processes. Yao et al. [47] found that a lncRNA, LINC00675, is upregulated in androgen-insensitive Pca cell lines and CRPC patients, and that LINC00675 can directly modulate the interaction of ganglion AR with dual microhomologous gene 2. Knockdown of LINC00675 gene can significantly inhibit Pca tumor formation and reduces enzalutamide resistance. These studies indicate that lncRNA LINC00675 is related to the resistance of enzalutamide through the modulation of AR signaling pathway and plays an important role in the development of CRPC.
MicroRNAs
MicroRNA(miRNA)s are a class of endogenous and small noncoding RNAs that downregulate gene expression in various ways, including translational repression, cleavage, and deadenylation of target mRNAs. miRNA expressions continue to alter while cells proceed through advanced cancer stages or phenotypes such as ADPC and CRPC. A list of miRNAs regulating the AR signaling pathway is still incomplete but would be of great interest for providing new targets for the development of small miRNA based prostate cancer therapeutics. Çağdaş Aktan et al. evaluated miR-625-5p and miR-874-3p upregulation in ADPC to CRPC transition in LNCaP-104R2 and LNCaP-Abl cell lines. Suppression of miR-625-5p or miR-874-3p resulted in decreased proliferation of CRPC cells [48]. Jani Silva et al. designed a study to evaluate the prognostic potential of nine miRNAs in the liquid biopsies (plasma) of mCRPC patients treated with second-generation androgen receptor axis-targeted (ARAT) agents, abiraterone acetate (AbA) and enzalutamide (ENZ). They identified that miR-16-5p, miR-145-5p, and miR-20a-5p was implicated in several processes, namely, cell cycle, proliferation, migration, survival, metabolism, and angiogenesis [49]. Aya Naiki-Ito et al. found that luteolin inhibits CRPC by AR-V7 suppression through miR-8080, highlighting luteolin and miR-8080 as promising therapeutic agents for this disease [50].
Circular RNA
Circular RNAs (circRNAs) as a non-coding form of RNA, widely expressed in many tissues with distinct function to influence development of several diseases including tumor progression. Despite their growing links to cancer, there has been limited characterization of circRNAs in metastatic CRPC, the major cause of prostate cancer mortality. Subing Cao et al. through the analysis of an exome-capture RNA seq dataset from 47 metastatic castration-resistant prostate cancer specimens and ribodepletion and RNase R RNA-sequencing of patient-derived xenografts (PDXs) and cell models, they identified 13 circRNAs generated from the key prostate cancer driver gene-androgen receptor (AR). They found that with greater resistance to exoribonuclease compared to the linear AR transcripts and detectability of AR circRNAs in patient plasma, these AR circRNAs may serve as surrogate circulating markers for AR/AR-variant expression and CRPC progression [51]. A study by Gang Wu et al. suggested that circRNA17 may function as suppressor to alter the Enz sensitivity and cell invasion in CRPC cells via altering the miR-181c-5p/ARv7 signaling and targeting this newly identified signaling may help in the development of a better therapy to further suppress the EnzR cell growth [52].
John Greene profile and demonstrated discrete circRNA expression patterns in an enzalutamide resistant cell line model of prostate cancer. He suggested that hsa_circ_0004870, through RBM39, may play a critical role in the development of enzalutamide resistance in CRPC [53].
Figure 2 shows that crosstalk between ncRNAs in AR regulatory network.
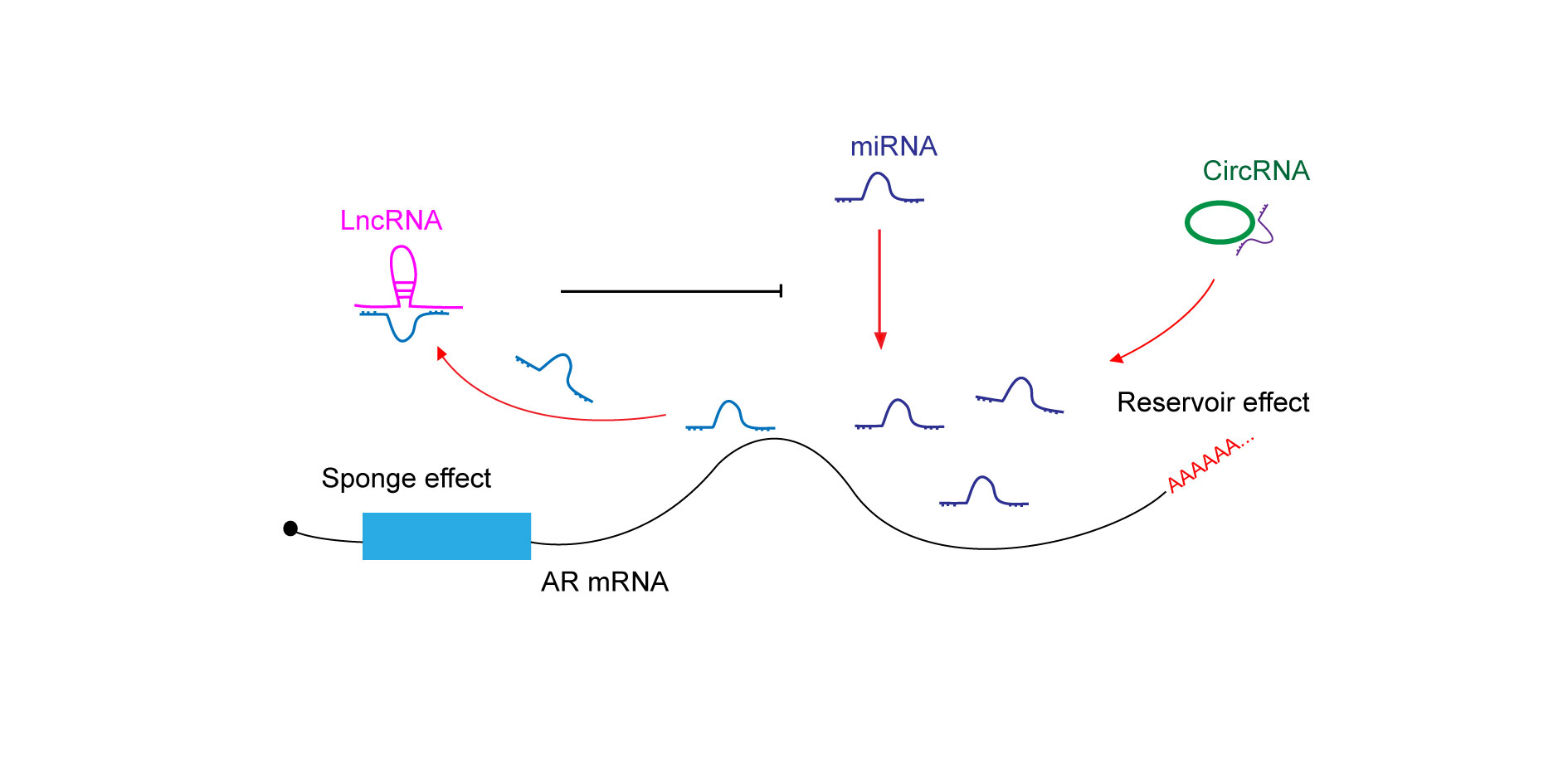 Figure 2.
Figure 2. Crosstalk between non-coding RNAs in AR regulatory network. LncRNA acts as a sponge to inhibit miRRNA targeted AR mRNA degradation. CircRNA binds to and stabilizes miRNA, enhancing miRNA targeted AR mRNA degradation.
Conclusions
In CRPC, AR is undoubtedly a hot spot. The traditional phosphatidylinositol 3-kinase/protein kinase B pathway, mitogen-activated protein kinase pathway and P53 signaling pathway play an important role in the development of Pca. With the transition of Pca emerging the CRPC stage, AR signaling pathway plays an important role in the development of CRPC. AR is overexpressed in the vast majority of CRPC, and the AR gene is amplified in 1/3 of cases. In addition, splicing variants and point mutation of AR may play a role in the progression of Pca, allowing AR to function independently of androgens. Studies have shown that overexpression of AR converts androgen-sensitive growth to non-androgen-dependent growth and hypersensitizes the AR signaling pathway. Thus, there is evidence that overexpression of AR is the main mechanism of partial castration resistance, and that overexpression of AR is partially explained by gene amplification. Comodulating factors can alter the transcriptional activity of AR. At present, there are different drugs to treat CRPC for different mechanisms, such as androgen biosynthesis inhibitor abiraterone, the second-generation AR antagonist Enzalutamide, chemotherapeutic drug docetaxel, etc. With the prolongation of ADT, AR signaling pathway promotes tumor resistance to drug resistance through various mechanisms. Preunderstanding of these mechanisms will help provide new ideas for drug development, such as the development of related drugs targeting the recruitment pathway of AR coactivators. With the in-depth understanding of AR signaling pathway, safer and more effective targeted drugs for CRPC in the future will certainly bring new hope for Pca patients.
Declaration
Acknowledgements
None.
Ethical policy
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study. Approval from institutional ethical committee was taken.
Availability of data and materials
All data generated or analysed during this study are included in this publication.
Author contributions
NC: Conception, design of study and manuscript preparation; MJL: Data collection and analysis and approval for the final version of the manuscript and funding supports.
Competing interests
The authors have no competing interest.
Funding
None.
References
-
Bergengren O, Pekala KR, Matsoukas K, Fainberg J, Mungovan SF, Bratt O, Bray F, Brawley O, Luckenbaugh AN, Mucci L et al: 2022 Update on Prostate Cancer Epidemiology and Risk Factors-A Systematic Review. Eur Urol 2023, 84(2): 191-206.
-
Liu J, Dong L, Zhu Y, Dong B, Sha J, Zhu HH, Pan J, Xue W: Prostate cancer treatment - China's perspective. Cancer Lett 2022, 550: 215927.
-
Shirley M: Relugolix: A Review in Advanced Prostate Cancer. Target Oncol 2023, 18(2): 295-302.
-
Menges D, Yebyo HG, Sivec-Muniz S, Haile SR, Barbier MC, Tomonaga Y, Schwenkglenks M, Puhan MA: Treatments for Metastatic Hormone-sensitive Prostate Cancer: Systematic Review, Network Meta-analysis, and Benefit-harm assessment. Eur Urol Oncol 2022, 5(6): 605-616.
-
Roy S, Saad F: Metastatic castrate-resistant prostate cancer: a new horizon beyond the androgen receptors. Curr Opin Support Palliat Care 2022, 16(4): 223-229.
-
Hadfield MJ, Lyall V, Holle LM, Dennison M: Updates in the Treatment of Non-Metastatic Castrate-Resistant Prostate Cancer: The Benefit of Second-Generation Androgen Receptor Antagonists. Ann Pharmacother 2023, 57(11): 1302-1311.
-
Mir N, Burke O, Yates S, Rajasekaran T, Chan J, Szmulewitz R, Kanesvaran R: Androgen receptor pathway inhibitors, prostate cancer, and older adults: a global Young International Society of Geriatric Oncology drug review. Ther Adv Med Oncol 2023, 15: 17588359221149887.
-
Senapati D, Sharma V, Rath SK, Rai U, Panigrahi N: Functional implications and therapeutic targeting of androgen response elements in prostate cancer. Biochimie 2023, https://doi.org/10.1016/j.biochi.2023.07.012. Epub ahead of print.
-
Wang W, Kong P, Feng K, Liu C, Gong X, Sun T, Duan X, Sang Y, Jiang Y, Li X et al: Exosomal miR-222-3p contributes to castration-resistant prostate cancer by activating mTOR signaling. Cancer Sci 2023, https://doi.org/10.1111/cas.15948. Epub ahead of print.
-
Militaru FC, Militaru V, Crisan N, Bocsan IC, Udrea AA, Catana A, Kutasi E, Militaru MS: Molecular basis and therapeutic targets in prostate cancer: A comprehensive review. Biomol Biomed 2023, 23(5): 760-771.
-
Edwards J, Krishna NS, Mukherjee R, Watters AD, Underwood MA, Bartlett JM: Amplification of the androgen receptor may not explain the development of androgen-independent prostate cancer. BJU Int 2001, 88(6): 633-637.
-
Van-Duyne G, Blair IA, Sprenger C, Moiseenkova-Bell V, Plymate S, Penning TM: The androgen receptor. Vitam Horm 2023, 123: 439-481.
-
Claessens F, Denayer S, Van Tilborgh N, Kerkhofs S, Helsen C, Haelens A: Diverse roles of androgen receptor (AR) domains in AR-mediated signaling. Nucl Recept Signal 2008, 6: e008.
-
Fernandes RC, Leach DA, Bevan CL: Epigenetic Coregulation of Androgen Receptor Signaling. Adv Exp Med Biol 2022, 1390: 277-293.
-
Wong HY, Demmers JA, Bezstarosti K, Grootegoed JA, Brinkmann AO: DNA dependent recruitment of DDX17 and other interacting proteins by the human androgen receptor. Biochim Biophys Acta 2009, 1794(2): 193-198.
-
Shafi AA, Yen AE, Weigel NL: Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol Ther 2013, 140(3): 223-238.
-
Jamroze A, Chatta G, Tang DG: Androgen receptor (AR) heterogeneity in prostate cancer and therapy resistance. Cancer Lett 2021, 518: 1-9.
-
Antonarakis ES, Chandhasin C, Osbourne E, Luo J, Sadar MD, Perabo F: Targeting the N-Terminal Domain of the Androgen Receptor: A New Approach for the Treatment of Advanced Prostate Cancer. Oncologist 2016, 21(12): 1427-1435.
-
Tan J, Sharief Y, Hamil KG, Gregory CW, Zang DY, Sar M, Gumerlock PH, deVere White RW, Pretlow TG, Harris SE et al: Dehydroepiandrosterone activates mutant androgen receptors expressed in the androgen-dependent human prostate cancer xenograft CWR22 and LNCaP cells. Mol Endocrinol 1997, 11(4): 450-459.
-
Krishnan AV, Zhao XY, Swami S, Brive L, Peehl DM, Ely KR, Feldman D: A glucocorticoid-responsive mutant androgen receptor exhibits unique ligand specificity: therapeutic implications for androgen-independent prostate cancer. Endocrinology 2002, 143(5): 1889-1900.
-
Chaturvedi AP, Dehm SM: Androgen Receptor Dependence. Adv Exp Med Biol 2019, 1210: 333-350.
-
Mizokami A, Namiki M: Reconsideration of progression to CRPC during androgen deprivation therapy. J Steroid Biochem Mol Biol 2015, 145: 164-171.
-
Qu X, Randhawa G, Friedman C, Kurland BF, Glaskova L, Coleman I, Mostaghel E, Higano CS, Porter C, Vessella R et al: A three-marker FISH panel detects more genetic aberrations of AR, PTEN and TMPRSS2/ERG in castration-resistant or metastatic prostate cancers than in primary prostate tumors. PLoS One 2013, 8(9): e74671.
-
Xia Y, Huang CC, Dittmar R, Du M, Wang Y, Liu H, Shenoy N, Wang L, Kohli M: Copy number variations in urine cell free DNA as biomarkers in advanced prostate cancer. Oncotarget 2016, 7(24): 35818-35831.
-
Wang J, Zhang W, Ji W, Liu X, Ouyang G, Xiao W: The von hippel-lindau protein suppresses androgen receptor activity. Mol Endocrinol 2014, 28(2): 239-248.
-
Shen M, Demers LK, Bailey SD, Labbe DP: To bind or not to bind: Cistromic reprogramming in prostate cancer. Front Oncol 2022, 12: 963007.
-
Zhou RW, Xu J, Martin TC, Zachem AL, He J, Ozturk S, Demircioglu D, Bansal A, Trotta AP, Giotti B et al: A local tumor microenvironment acquired super-enhancer induces an oncogenic driver in colorectal carcinoma. Nat Commun 2022, 13(1): 6041.
-
Mao Y, Yang G, Li Y, Liang G, Xu W, Hu M: Advances in the Current Understanding of the Mechanisms Governing the Acquisition of Castration-Resistant Prostate Cancer. Cancers (Basel) 2022, 14(15).
-
Sumiyoshi T, Mizuno K, Yamasaki T, Miyazaki Y, Makino Y, Okasho K, Li X, Utsunomiya N, Goto T, Kobayashi T et al: Clinical utility of androgen receptor gene aberrations in circulating cell-free DNA as a biomarker for treatment of castration-resistant prostate cancer. Sci Rep 2019, 9(1): 4030.
-
Cato L, Shomali M: AR Structural Variants and Prostate Cancer. Adv Exp Med Biol 2022, 1390: 195-211.
-
Lu C, Brown LC, Antonarakis ES, Armstrong AJ, Luo J: Androgen receptor variant-driven prostate cancer II: advances in laboratory investigations. Prostate Cancer Prostatic Dis 2020, 23(3): 381-397.
-
Yuan S, He SH, Li LY, Xi S, Weng H, Zhang JH, Wang DQ, Guo MM, Zhang H, Wang SY et al: A potassium-chloride co-transporter promotes tumor progression and castration resistance of prostate cancer through m(6)A reader YTHDC1. Cell Death Dis 2023, 14(1): 7.
-
Takayama KI, Fujimura T, Suzuki Y, Inoue S: Identification of long non-coding RNAs in advanced prostate cancer associated with androgen receptor splicing factors. Commun Biol 2020, 3(1): 393.
-
Takayama KI: Splicing Factors Have an Essential Role in Prostate Cancer Progression and Androgen Receptor Signaling. Biomolecules 2019, 9(4).
-
Reichert ZR, Kasputis T, Nallandhighal S, Abusamra SM, Kasputis A, Haruray S, Wang Y, Williams S, Singhal U, Alva A et al: Multigene Profiling of Circulating Tumor Cells (CTCs) for Prognostic Assessment in Treatment-Naive Metastatic Hormone-Sensitive Prostate Cancer (mHSPC). Int J Mol Sci 2021, 23(1).
-
Wang J, Park KS, Yu X, Gong W, Earp HS, Wang GG, Jin J, Cai L: A cryptic transactivation domain of EZH2 binds AR and AR's splice variant, promoting oncogene activation and tumorous transformation. Nucleic Acids Res 2022, 50(19): 10929-10946.
-
Howard N, Clementino M, Kim D, Wang L, Verma A, Shi X, Zhang Z, DiPaola RS: New developments in mechanisms of prostate cancer progression. Semin Cancer Biol 2019, 57: 111-116.
-
Mohsenzadegan M, Razmi M, Vafaei S, Abolhasani M, Madjd Z, Saeednejad Zanjani L, Sharifi L: Co-expression of cancer-testis antigens of MAGE-A6 and MAGE-A11 is associated with tumor aggressiveness in patients with bladder cancer. Sci Rep 2022, 12(1): 599.
-
Su S, Parris AB, Grossman G, Mohler JL, Wang Z, Wilson EM: Up-Regulation of Follistatin-Like 1 By the Androgen Receptor and Melanoma Antigen-A11 in Prostate Cancer. Prostate 2017, 77(5): 505-516.
-
Biron E, Bedard F: Recent progress in the development of protein-protein interaction inhibitors targeting androgen receptor-coactivator binding in prostate cancer. J Steroid Biochem Mol Biol 2016, 161: 36-44.
-
Santosh B, Varshney A, Yadava PK: Non-coding RNAs: biological functions and applications. Cell Biochem Funct 2015, 33(1): 14-22.
-
Wu X, Ruan L, Yang Y, Mei Q: Identification of crucial regulatory relationships between long non-coding RNAs and protein-coding genes in lung squamous cell carcinoma. Mol Cell Probes 2016, 30(3): 146-152.
-
Dong R, Liu J, Sun W, Ping W: Comprehensive Analysis of Aberrantly Expressed Profiles of lncRNAs and miRNAs with Associated ceRNA Network in Lung Adenocarcinoma and Lung Squamous Cell Carcinoma. Pathol Oncol Res 2020, 26(3):1935-1945.
-
Misawa A, Takayama KI, Inoue S: Long non-coding RNAs and prostate cancer. Cancer Sci 2017, 108(11): 2107-2114.
-
Parolia A, Venalainen E, Xue H, Mather R, Lin D, Wu R, Pucci P, Rogalski J, Evans JR, Feng F et al: The long noncoding RNA HORAS5 mediates castration-resistant prostate cancer survival by activating the androgen receptor transcriptional program. Mol Oncol 2019, 13(5): 1121-1136.
-
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P et al: The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017, 45(D1): D362-D368.
-
Yao M, Shi X, Li Y, Xiao Y, Butler W, Huang Y, Du L, Wu T, Bian X, Shi G et al: LINC00675 activates androgen receptor axis signaling pathway to promote castration-resistant prostate cancer progression. Cell Death Dis 2020, 11(8): 638.
-
Aktan C, Cal C, Kaymaz B, Selvi Gunel N, Kipcak S, Ozel B, Gunduz C, Sahin Kucukaslan A, Aygunes Jafari D, Kosova B: Functional roles of miR-625-5p and miR-874-3p in the progression of castration resistant prostate cancer. Life Sci 2022, 301: 120603.
-
Silva J, Tavares V, Afonso A, Garcia J, Cerqueira F, Medeiros R: Plasmatic MicroRNAs and Treatment Outcomes of Patients with Metastatic Castration-Resistant Prostate Cancer: A Hospital-Based Cohort Study and In Silico Analysis. Int J Mol Sci 2023, 24(10).
-
Naiki-Ito A, Naiki T, Kato H, Iida K, Etani T, Nagayasu Y, Suzuki S, Yamashita Y, Inaguma S, Onishi M et al: Recruitment of miR-8080 by luteolin inhibits androgen receptor splice variant 7 expression in castration-resistant prostate cancer. Carcinogenesis 2020, 41(8): 1145-1157.
-
Cao S, Ma T, Ungerleider N, Roberts C, Kobelski M, Jin L, Concha M, Wang X, Baddoo M, Nguyen HM et al: Circular RNAs add diversity to androgen receptor isoform repertoire in castration-resistant prostate cancer. Oncogene 2019, 38(45): 7060-7072.
-
Wu G, Sun Y, Xiang Z, Wang K, Liu B, Xiao G, Niu Y, Wu D, Chang C: Preclinical study using circular RNA 17 and micro RNA 181c-5p to suppress the enzalutamide-resistant prostate cancer progression. Cell Death Dis 2019, 10(2): 37.
-
Greene J, Baird AM, Casey O, Brady L, Blackshields G, Lim M, O'Brien O, Gray SG, McDermott R, Finn SP: Circular RNAs are differentially expressed in prostate cancer and are potentially associated with resistance to enzalutamide. Sci Rep 2019, 9(1): 10739.
Cite this article: Cui Na, Li MJ: Research Progress on the Mechanism of Androgen Receptor Signaling Pathway in Castration-Resistant Prostate Cancer. Ann Urol Oncol 2023, 6(4): 132-138. https://doi.org/10.32948/auo.2023.11.09
 Figure 1. Schematic structure of human androgen receptor (AR) and AR splice variant 7 (AR-V7) and 567(AR567es). (a) The full-length AR; (b) AR-V7 (also named AR3) encodes a protein with exons 1-3 and a terminal cryptic exon (CE3); (c) AR567es encodes a protein comprised of exons 1-4, and because of a frame-shift due to loss of exons 5-7, exon 8 has a stop codon generated after the first 10 amino acids resulting in a shortened exon 8.
Figure 1. Schematic structure of human androgen receptor (AR) and AR splice variant 7 (AR-V7) and 567(AR567es). (a) The full-length AR; (b) AR-V7 (also named AR3) encodes a protein with exons 1-3 and a terminal cryptic exon (CE3); (c) AR567es encodes a protein comprised of exons 1-4, and because of a frame-shift due to loss of exons 5-7, exon 8 has a stop codon generated after the first 10 amino acids resulting in a shortened exon 8.
 Figure 2. Crosstalk between non-coding RNAs in AR regulatory network. LncRNA acts as a sponge to inhibit miRRNA targeted AR mRNA degradation. CircRNA binds to and stabilizes miRNA, enhancing miRNA targeted AR mRNA degradation.
Figure 2. Crosstalk between non-coding RNAs in AR regulatory network. LncRNA acts as a sponge to inhibit miRRNA targeted AR mRNA degradation. CircRNA binds to and stabilizes miRNA, enhancing miRNA targeted AR mRNA degradation.
 Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
 Submit Manuscript
Submit Manuscript