Review Article | Open Access
The Role of Urinary Extracellular Vesicles in Kidney Cancer: Diagnostic and Therapeutic Potential
Kirk Gallego1
1Universidad de Montemorelos, Health Sciences Department, Montemorelos, N.L., Mexico.
Correspondence: Kirk Gallego (Universidad de Montemorelos, Health Sciences Department, Montemorelos, N.L., Mexico; Email: KirkGallego2@outlook.com).
Annals of Urologic Oncology 2024, 7(4): 158-167. https://doi.org/10.32948/auo.2024.10.25
Received: 15 Oct 2024 | Accepted: 06 Nov 2024 | Published online: 17 Nov 2024
Key words extracellular vesicles, urinary extracellular vesicles, renal cell cancer, urine, biomarker
Extracellular vesicles (EVs) are membrane-bound particles composed of a lipid bilayer that carries a variety of biological factors, including nucleic acids, proteins, metabolites, and lipids. EVs are available in different forms and can generally be divided into larger and smaller groups. They consisted of exosomes, large oncosomes, apoptotic bodies, migrasomes, ectosomes (shedding microvesicles), and exosomes. Large extracellular vesicles are generally identified with diameters greater than 200 nm. This category includes different types like large oncosomes, apoptotic bodies, migrasomes, and ectosomes/shedding microvesicles (Figure 1). Conversely, small extracellular vesicles consist of vesicles that have diameters smaller than 200 nm. These consist of exosomes, exomeres, and a particular group of ectosomes/shedding microvesicles (Figure 2) [3-6]. Body fluids, such as blood, plasma, serum, and urine, along with a tissue medium for culture, serve as valuable sources of EVs [7]. A recent study indicated that EVs can be obtained from donor cells and transferred to recipient cells, thereby serving as a novel method of intercellular communication [8]. Moreover, EVs possess biomarkers that may be used to identify future metastatic sites. Therapeutic strategies may target EVs, inhibit specific organ uptake, target EV-induced alterations in possible metastatic sites, and utilize EVs as a drug delivery channel [9]. The prospective function of EVs in identifying and treating diseases with optimal accuracy has generated considerable research interest. This review summarizes the current research examining the potential utility of urinary extracellular vesicle (uEVs) cargo as biomarkers for prognosis, diagnosis, and medical therapy in kidney cancer.
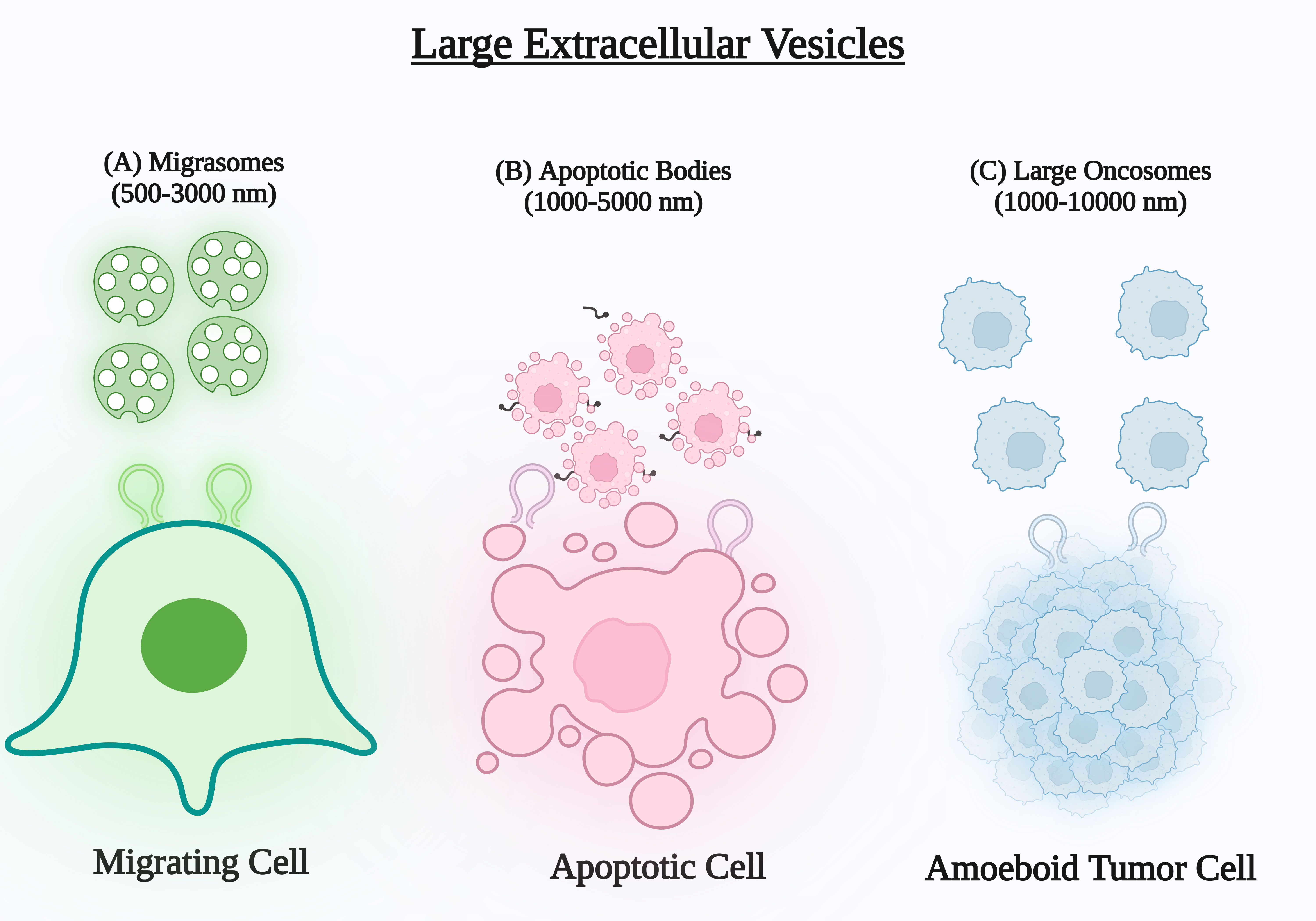 Figure 1 (A) Migrasomes are produced during cell migration at the ends or intersections of retraction fibres. They are then released into the extracellular matrix when the fibres separate. (B) Apoptotic bodies are discharged from the cell by membrane projections or blebs formed during apoptosis and released into the extracellular matrix. (C) Oncosomes are frequently produced from protrusions on the outer layer of amoeboid tumour cells and have been discovered to convey cancerous substances.
Figure 1 (A) Migrasomes are produced during cell migration at the ends or intersections of retraction fibres. They are then released into the extracellular matrix when the fibres separate. (B) Apoptotic bodies are discharged from the cell by membrane projections or blebs formed during apoptosis and released into the extracellular matrix. (C) Oncosomes are frequently produced from protrusions on the outer layer of amoeboid tumour cells and have been discovered to convey cancerous substances.
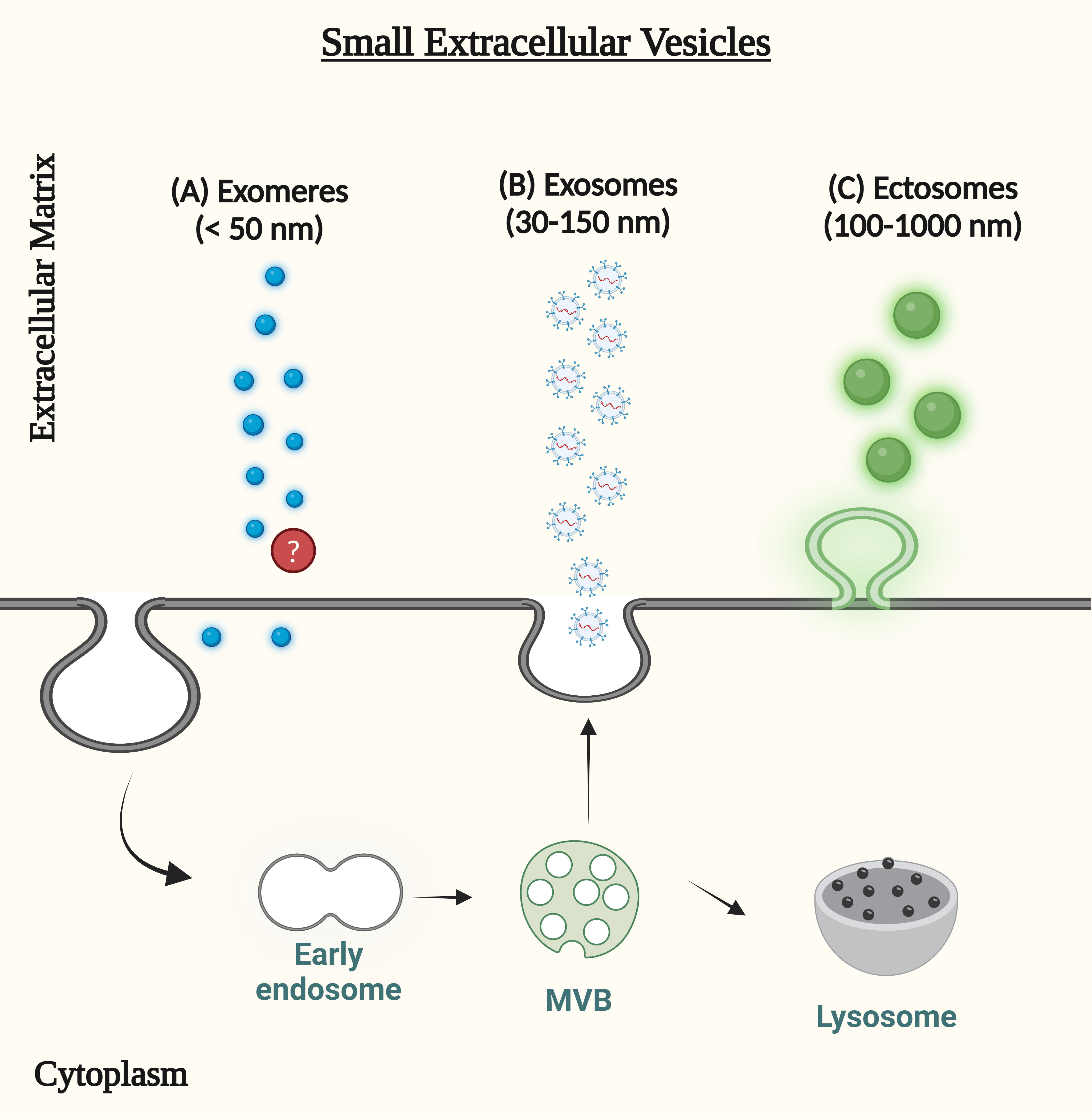 Figure 2 (A) Exomeres are extracellular vesicle-like non-membranous nanoparticles, but both their origin and the precise mechanisms underlying their production are still unclear (B) Endocytic vesicles merge with early endosomes to generate exosomes. Membrane budding creates intraluminal vesicles in the beginning endosomes. By fusing with the cell membrane, lumen vesicles can be destroyed by lysosomes or released into the extracellular environment. Released vesicles are called exosomes. (C) The outward protrusion of the plasma membrane releases ectosomes, often referred to as shedding microvesicles, into the extracellular matrix.
Figure 2 (A) Exomeres are extracellular vesicle-like non-membranous nanoparticles, but both their origin and the precise mechanisms underlying their production are still unclear (B) Endocytic vesicles merge with early endosomes to generate exosomes. Membrane budding creates intraluminal vesicles in the beginning endosomes. By fusing with the cell membrane, lumen vesicles can be destroyed by lysosomes or released into the extracellular environment. Released vesicles are called exosomes. (C) The outward protrusion of the plasma membrane releases ectosomes, often referred to as shedding microvesicles, into the extracellular matrix.
Physiological functions of uEVs
Growing evidence suggests that EVs excreted in urine can be internalized by other cells, hence influencing their function, indicating the existence of intra-nephron interaction along the urinary lumen [14]. Studies using electron microscopy have shown that cilia in vitro allow proximal tubular epithelial cells to internalize EVs [12]. Additionally, in vitro investigations have demonstrated that tubular cells may internalize EVs produced from collecting ducts, thereby transmitting aquaporin 2 (AQP2) [15]. By inducing a profibrotic phenotype in cultured tubular epithelial cells, podocyte-derived EVs may have discovered a new method of glomerular-tubular communication [16, 17].
Furthermore, uEVs are likely to play a major role in elimination because they accumulate in the bladder and are subsequently expelled through urine, which is a heterogeneous mixture of uEVs. The major route of EVs elimination, including circulating EVs, is not yet known to be excreted through urine; rather, it is unclear whether EVs from the urinary tract are expelled from urine. Research on the physiological roles of uEVs is still in its early stages of development.
uEVs isolation methods
Innovation in the scientific and technological fields has driven the advancement of various EV isolation techniques. However, there has yet to be an ideal approach. Consequently, when designing an EV study, it is imperative to select the isolation method according to the intended downstream application (protein or nucleic acid isolation, biomarker finding, or functional assays) and the biological fluid from which the EVs will be collected (cultured cell media, urine, serum, plasma) [18-20]. The isolation of uEVs requires consideration of numerous practical factors[13, 21]. In research using urine extracellular vesicles, upholding ideal storage conditions for urine samples to avert proteolysis is crucial. Storage at 80°C, as opposed to at 4 °C or 20°C, is advantageous for avoiding degradation. However, the use of freshly processed urine is the most effective approach [22].
uEVs have been isolated using various methods, including ultracentrifugation, chemical precipitation, size-exclusion chromatography, and ultrafiltration technology [23]. Each technique utilises specific biophysical or biochemical characteristics of uEVs, including size, mass weight, structure, charge, and surface proteins, to facilitate their isolation. The two most common types of isolation procedures are ultracentrifugation (UC) and density-gradient ultracentrifugation (dUC) [24]. However, under pathological conditions, the resultant uEVs pellet is contaminated with prevalent urine proteins, including Tamm-Horsfall protein (THP, or uromodulin) and albumin. THP is derived from a glycosylphosphatidylinositol-linked protein located in the apical membrane of the thick ascending limb of the loop of Henle and is released into the urine through proteolytic processing [25]. THP generate extensive fiber channels that can restrict uEVs in urine and affect filtration mechanisms [26]. Therefore, methods to reduce or eradicate THP in the urine before uEVs isolation are necessary to improve the final production. Advanced ultracentrifugation is the predominant technique employed [27].
Table 1 outlines the various techniques for isolating uEVs.
Characterization methods
The widespread interest and complexity of uEVs have led to the invention and application of numerous approaches for their characterization. The currently used protocols and commercial kits that assert the isolation or purification of exosomes or EVs to a high standard are not capable of completely separating EVs from non-EV entities [38], making EV characterization a challenging task. Non-EV entities, for example, the argonaut 2 protein complex and lipoproteins, also contain components found in EVs [39, 40]. Furthermore, the results can be difficult to interpret because of the variety of methods and the composition of recovered EVs, which are influenced by factors such as experimental system variability, investigator expertise, and the apparatus employed. To ensure that biomarkers are linked to EVs and are not contaminated, EV detection and characterization should be evaluated using a variety of complementary approaches, as per the MISEV2018 guidelines [18].
Unfortunately, no single technique can accurately describe the morphology, size, amount, and content of uEVs. Modern methods employed for the morphological characterization of EVs encompass transmission electron microscopy (TEM), cryogenic electron microscopy (cryo-EM), atomic force microscopy (AFM), and super-resolution fluorescence microscopy, with TEM being the most prevalent [41]. These methods are not always compatible with one another or can provide images of equal quality, because they provide distinct details regarding the structure and size distribution of uEVs. The techniques employed to quantify the size distribution and count of uEVs include nanoparticle tracking analysis (NTA) and tunable resistive pulse sensing (TRPS) [42, 43]. Based on Brownian motion, NTA offers the concentration and size distribution of uEVs particles within a certain detection range; however, it cannot rule out non-EV entities. Therefore, the particle count obtained from NTA may have been inflated. TRPS is a more precise way for measuring the particle size, amount, and surface charge [44, 45]. The concentration and size distribution of uEVs in a sample can be estimated by evaluating a large number of trajectories. Nevertheless, this technique is limited by the brief measured trajectories of in-focus and out-of-focus vesicles and particles, which may result in an inaccurate estimation of the particle concentration [46].
Finally, to enhance the integrity of uEVs preparation, co-isolated components must be analyzed (e.g., THP in urine) [18]. Although immunoblotting, TEM and NTA are the most common methods for characterizing EVs [47], there is a vast array of approaches that are currently being developed and will contribute to better EV characterization in the future. One way to achieve good EV characterization is to combine some of the strategies that have been mentioned [48].
|
Table 1. Different methods for isolating uEVs with pros and cons of each method. |
||||
|
Techniques |
Methods of Isolations |
Pros |
Cons |
Ref |
|
Ultracentrifugation
|
1. Progressive ultracentrifugation 2. Double-cushion ultracentrifugation 3. Sucrose gradient ultracentrifugation 4. Ultracentrifugation size exclusion chromatography |
1. Results are reproducible, demonstrating many intact proteins and nucleic acids. 2. Reduced contamination of highly abundant proteins. |
1. Processing a single sample requires 5-7 hours, is susceptible to contamination by highly abundant proteins, and requires costly machinery. 2. Extended processing duration; challenging separation methods; costly apparatus |
[28-30] |
|
Filtration |
1. Nanomembrane filtration 2. Micromembrane filtration |
Reduced processing duration (0.5-2 hours); several samples can be simultaneously processed; cost-effective; applicable in clinical environments. |
Risk of membrane blockage, specimen loss, and contamination from abundant proteins. |
[31, 32] |
|
Precipitation
|
Precipitation by ExoQuick-TC |
Reduced processing time (0.5–2 hours); cost-effective; yields RNA in its intact form; applicable to therapeutic settings |
Protein purity is low; the protocol has been modified. |
[33] |
|
Hydrostatic dialysis |
Hydrostatic filtration dialysis
|
Appropriate for any downstream analysis; low cost, basic system; effective preparation and concentration for biobanking reasons. |
Low protein purity compared to ultracentrifugation, but acceptable; THP contamination present. Compared to ultracentrifugation, large vesicles (>500 nm) were minimal, small EVs (60-140 nm) were rare, and EV-like particles (<40 nm) were more prevalent. |
[34, 35] |
|
Acoustic trapping |
Polystyrene beads model |
Quick, automated, suitable with low volumes, and resilient; does not affect trapped vesicle integrity or miRNA content
|
Device parallelization may not be possible if an amplifier drives the piezo. |
[36, 37] |
The diagnosis of RCC is frequently incidental, as many cases remain asymptomatic until advanced stages of the disease. Consequently, the diagnosis of RCC is frequently postponed until the disease has progressed, with 30% of patients presenting with metastasis at diagnosis and an additional 30% developing metastasis during the disease's progression [54]. The gold standard solution for the management of localized kidney cancer is kidney surgery. Such procedures include radical nephrectomy, which involves the removal of the entire kidney, and partial nephrectomy, which involves the removal of only the section of the kidney that is malignant [55]. Additional methods for treating kidney tumor include radiation therapy, chemotherapy, targeted therapies, cryoablation, radiofrequency ablation, and microwave ablation [56, 57]. Furthermore, engineered EVs are promising carriers for RCC to enhance our understanding of targeted therapies.
Multiple ground-breaking investigations have demonstrated the potential of EVs in RCC diagnosis. CA9, CD70, and CD147, notable markers expressed in ccRCC tumour tissues, are also found in secreted EVs. These proteins can be used as reliable biomarkers for tumor-specific, noninvasive detection approaches since their expression in EVs proves that they originate from the main cells of the kidney [58]. The engineered MSC-derived EVs that were designed with TRAIL (TNF-related apoptosis inducing ligand) shown a substantial effect on TRAIL-resistant renal carcinoma cell lines, such as RCC10 and HA7-RCC examples [59]. MSC-derived EVs have a moderate effect on renal cancer by promoting apoptosis and inhibiting proliferation. RCC is an important concern owing to its high metastatic rate, death rate, increased incidence, and resistance to treatment. It becomes difficult to diagnose solid tumours when there are unusual tumour cell patterns or insufficient tissue samples [60, 61].
EVs originating from ccRCC, papillary RCC (pRCC), and benign renal cell lines exhibit distinct signatures, enabling differentiation not only between RCC subtypes but also between RCC and benign renal cells [62]. EVs generated by ccRCC and pRCC, respectively, are specifically enriched in exosomal proteins, while EVs of benign renal cells, but not ccRCC, include distinct exosomal mRNA of EPCAM, PRKCZ, PXDN, CXADR, EPS8L1, HOXA7, LAD1, MYO1D, ROCK2, and SLC35A3 [63]. Conversely, the epithelial tumour cell marker EpCAM is broadly expressed in both normal tubular and ccRCC samples. Furthermore, CDH2, COL7A1, FGFR2, BMPR1B, HDHD3, ICAM1, KIAA1462, and PFKFB4 mRNA are only present in EVs that are produced from ccRCCs [58, 63].
In addition to proteins and mRNAs, EVs are significantly enriched with non-coding RNAs, such as microRNAs (miRNAs), circular RNAs, and long non-coding RNAs (lncRNAs). Various miRNAs transported by EVs exhibit unique expression profiles when comparing patients with RCC to individuals with good health. Grange and his fellows identified that within the CD105+ microvesicles, there was a notable upregulation of 24 miRNAs, such as miR-200c and miR-650, alongside a downregulation of 33 miRNAs, including miR-100 and miR-29 [64]. Furthermore, Zhang and his team observed that serum samples from patients with ccRCC displayed significantly heightened levels of exosomal miR-210 and miR-1233 in comparison to healthy controls, with these levels markedly diminishing following nephrectomy [65]. Therefore, the detection of exosomal miR-210 and miR-1233 in serum may prove to be a significant marker in the diagnosis and ongoing assessment of patients with ccRCC, especially in the context of liquid biopsies. Wang and his workfellows carried out research investigating serum exosomal miR-210, noting its upregulation in ccRCC, especially among patients presenting with advanced tumour stages, elevated Fuhrman grades, and the presence of metastases [66]. Fujii and colleagues performed an investigation into the serum levels of exosomal miR-224, emphasising its negative prognostic significance in patients with ccRCC. Their findings further elucidated the significant potential of exosomal miR 224 as a predictive biomarker for the detection of microinvasion or tumor metastasis subsequent to nephrectomy in patients with ccRCC [67].
Exosomal miR-21-5p derived from M2 macrophages has been linked to pro-metastatic effects in RCC via the activation of the PTEN/Akt pathway. A recent research revealed that the suppression of miR-21-5p within M2 exosomes led to a reduction in the metastatic capabilities of RCC cells [68]. Moreover, research has demonstrated that serum exosomes exhibit a markedly elevated gamma-glutamyl transferase (GGT) activity in individuals with advanced RCC, as well as those presenting with metastatic disease and microvascular invasion [69]. Hence, integrating exosomal GGT with standard diagnostic procedures may improve ccRCC diagnosis. Recent research has found higher exosomal MYO15A levels in the serum of patients with ccRCC, indicating a poorer prognosis and potential as a diagnostic target [70].
LncRNAs are RNA molecules that exceed 200 nucleotides in length and play a crucial role in regulating cellular processes, including transcription and protein translation, through their interactions with proteins, mRNAs, or miRNAs. They show certain expression patterns in tumor cells, which could make them useful for cancer diagnosis [71, 72]. Studies have shown that the transfer of lncARSR via exosomes enhances the expression of AXL and c-MET in RCC cells by competitively interacting with miR-34/miR-449, thus playing a role in the emergence of resistance to sunitinib [73, 74]. Therefore, EVs are the most efficient means for transferring drug resistance in advanced RCC. These lncRNAs have the potential to be used as treatment strategies for chemotherapy resistance and prognostic indicators. Although there is potential, additional research is required to broaden and apply the use of EVs to identify new RCC biomarkers.
Tumor-derived EVs play a significant role in influencing the tumor’s microenvironment, thereby sustaining, and promoting the growth of cancer [64]. Several specific markers of RCC were found in one of the first studies to compare the proteomic profiles of uEVs from patients with RCC and healthy individuals. These included decreased expression of neprilysin, extracellular matrix metalloproteinase inducer (EMMPRIN; also known as basigin), dipeptidase 1, syntenin 1, and AQP1, and increased expression of metalloproteinase 9 (MMP9), ceruloplasmin, podocalyxin, carbonic anhydrase 9 (CAIX), and Dickkopf-related protein 4 (DKK4) [79]. A clinical study is currently underway to identify uEVs expressing CAIX in conjunction with exosomal marker CD9 in the urine of patients with ccRCC using electron microscopy [80].
Research has also explored uEVs miRNAs as potential biomarkers of RCC. It has been observed that individuals with ccRCC can be distinguished from healthy individuals by the presence of elevated levels of miR-126-3p in conjunction with miR-449a or miR-34b-5p in the cargo of uEVs [81]. Another research study indicated the elevation of miR-204-5p in urinary exosomes from patients with Xp11 translocation RCC, an extremely uncommon sporadic pediatric renal cancer, implying that this increase may serve as a valuable diagnostic for early diagnosis [82]. Metastatic RCC and poor survival rates are strongly linked to decreased miR-126 expression, which is induced by lncRNA DUXAP8 [83].
Kuczler et al. conducted a comparison analysis of exosomal mRNA in urine and tissue models from patients with RCC. Exosomal mRNA transcripts of ALOX5, RBL2, VEGFA, and TLK2 are particularly detected in the tissue and uEVs of patients with ccRCC [84]. In addition, patients with early stage ccRCC showed substantial downregulation of uEV-derived mRNA transcripts of NME2, AAMP, CAPNS1, VAMP8, and MYL12B [85]. miR-224-5p is highly elevated in both uEVs and tissues of patients with RCC compared with healthy controls. miR-224-5p stabilizes the expression of PD-L1 (programmed cell death protein 1) by directly inhibiting cyclin D1 (CCND1). This study clarified the mechanism by which miR-224-5p enhances resistance to T cell-dependent toxicity and metastasis by EV transmission among RCC cells [86].
Boccio et al. identified possible lipid biomarkers for RCC through the analysis of uEVs from RCC patients. These cancer-derived extracellular vesicles possess a distinctive lipidome comprising phosphatidylinositol phosphates (PIP), lysophospholipids (Lyso), phosphatidylethanolamines (PE), phosphatidylcholines (PC), mono-, di-, and triglycerides (MG/DG/TG), phosphatidic acids (PA), gangliosides (GL), and prostanoids (Pn) [87]. In conclusion, uEVs have demonstrated potential as the best option for minimally invasive, extremely sensitive, and unique renal cancer screening and treatment. Figure 3 illustrates the methods for processing uEV-derived miRNA and protein indicators for renal malignancies.
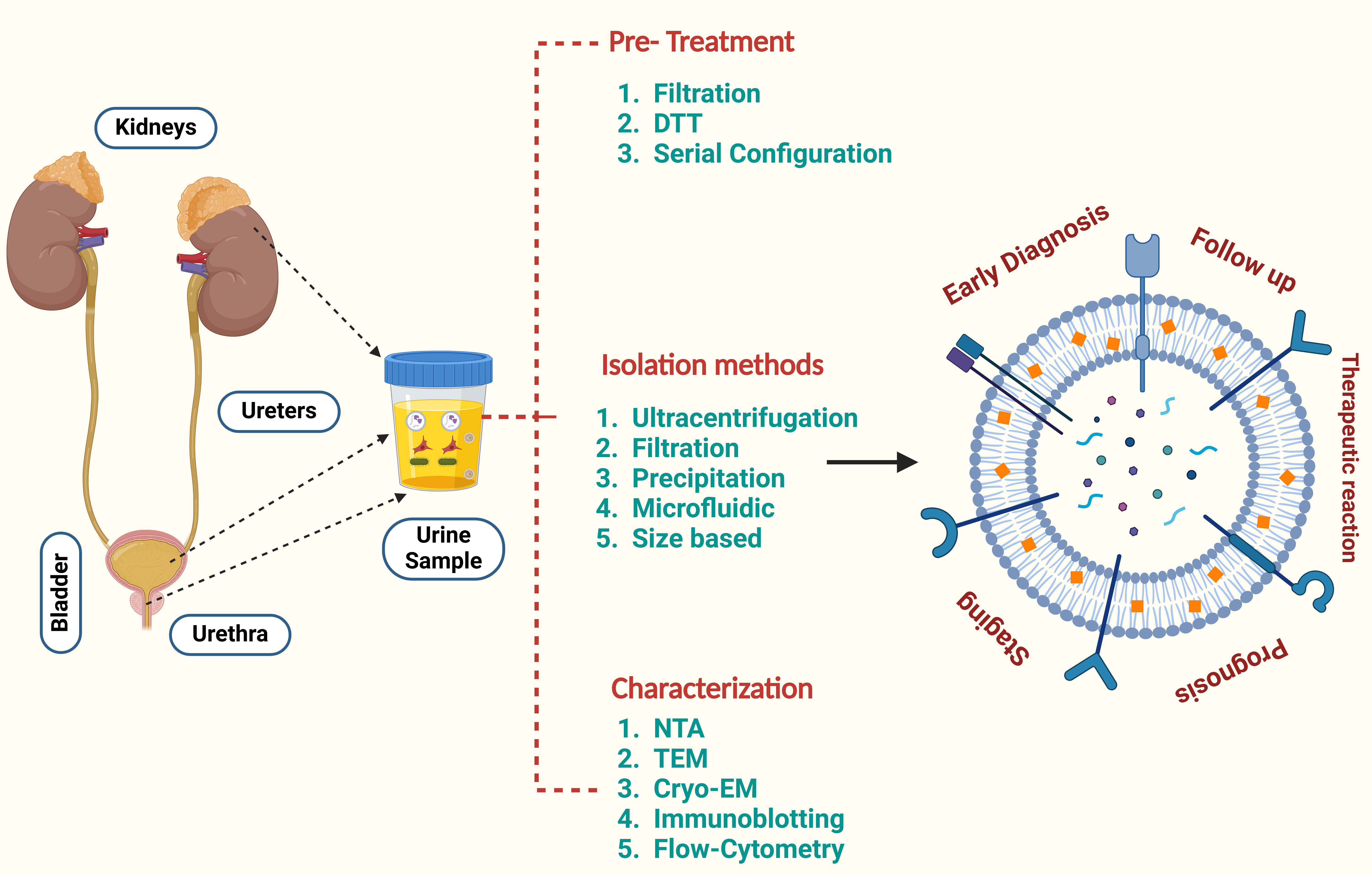 Figure 3. An overview of the processing stages for renal cancer biomarkers derived from uEVs, including miRNA and protein biomarkers.
Figure 3. An overview of the processing stages for renal cancer biomarkers derived from uEVs, including miRNA and protein biomarkers.At present, research on uEVs is predominantly limited to preclinical studies and initial clinical trials, which face obstacles in the translation of experimental results into therapeutic applications. Additional thorough studies and clinical trials are necessary to enable the use of uEVs in clinical settings. Future studies should include a larger number of samples and a variety of tissue types, utilizing prospective study designs that provide stronger evidence and more reliable medical information to facilitate clinical translation. Moreover, the analysis of uEVs in relation to RCC has been somewhat limited, and none of the identified molecules have been consistently validated in various studies. This highlights the need for further prospective clinical trials to identify reliable biomarkers.
In conclusion, uEVs are an appropriate medium for diagnosis because of their ease of collection and ability to represent the pathological conditions of the kidneys. Rapid urinary screening of predicted EV biomarkers could assist clinicians in diagnosis and facilitate the prompt selection of suitable therapies. Ultimately, its applications may have significant academic and commercial implications.
None.
Ethical policy
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study. Approval from institutional ethical committee was taken.
Availability of data and materials
All data generated or analysed during this study are included in this publication.
Author contributions
KG searched academic literature, wrote the draft manuscript, made the figures and submitted the final manuscript.
Competing interests
Authors report no conflict of interest.
Funding
None.
- Organization WH: WHO global survey on the inclusion of cancer care in health-benefit packages, 2020-2021. Epub ahead of print.: World Health Organization; 2024.
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F: Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021, 71(3): 209-249.
- Kalra H, Drummen GP, Mathivanan S: Focus on extracellular vesicles: introducing the next small big thing. Int J Mol Sci 2016, 17(2): 170.
- Chuo ST, Chien JC, Lai CP: Imaging extracellular vesicles: current and emerging methods. J Biomed Sci 2018, 25(1): 91.
- Ahmed W, Kuniyan MS, Jawed AM, Chen L: Engineered extracellular vesicles for drug delivery in therapy of stroke. Pharmaceutics 2023, 15(9): 2173.
- Ahmed W, Huang S, Chen L: Engineered exosomes derived from stem cells: a new brain-targeted strategy. Expert Opin Drug Deliv 2024, 21(1): 91-110.
- Zieren RC, Dong L, Pierorazio PM, Pienta KJ, de Reijke TM, Amend SR: Extracellular vesicle isolation from human renal cancer tissue. Med Oncol 2020, 37(4): 28.
- Butz H, Nofech-Mozes R, Ding Q, Khella HW, Szabó PM, Jewett M, Finelli A, Lee J, Ordon M, Stewart R, et al: Exosomal microRNAs are diagnostic biomarkers and can mediate cell–cell communication in renal cell carcinoma. Eur Urol Focus 2016, 2(2): 210-218.
- Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, Zambirinis CP, Rodrigues G, Molina H, Heissel S, et al: Extracellular vesicle and particle biomarkers define multiple human cancers. Cell 2020, 182(4): 1044-1061. e1018.
- Pisitkun T, Shen R-F, Knepper MA: Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA 2004, 101(36): 13368-13373.
- Kanno K, Sasaki S, Hirata Y, Ishikawa S-e, Fushimi K, Nakanishi S, Bichet DG, Marumo F: Urinary excretion of aquaporin-2 in patients with diabetes insipidus. N Engl J Med 1995, 332(23): 1540-1545.
- Hogan MC, Manganelli L, Woollard JR, Masyuk AI, Masyuk TV, Tammachote R, Huang BQ, Leontovich AA, Beito TG, Madden BJ, et al: Characterization of PKD protein-positive exosome-like vesicles. J Am Soc Nephrol 2009, 20(2): 278-288.
- Dear JW, Street JM, Bailey MA: Urinary exosomes: a reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics 2013, 13(10-11): 1572-1580.
- Gildea JJ, Seaton JE, Victor KG, Reyes CM, Wang DB, Pettigrew AC, Courtner CE, Shah N, Tran HT, Van Sciver RE, et al: Exosomal transfer from human renal proximal tubule cells to distal tubule and collecting duct cells. Clin Biochem 2014, 47(15): 89-94.
- Street JM, Birkhoff W, Menzies RI, Webb DJ, Bailey MA, Dear JW: Exosomal transmission of functional aquaporin 2 in kidney cortical collecting duct cells. J Physiol 2011, 589(24): 6119-6127.
- Munkonda MN, Akbari S, Landry C, Sun S, Xiao F, Turner M, Holterman CE, Nasrallah R, Hébert RL, Kennedy CR: Podocyte-derived microparticles promote proximal tubule fibrotic signaling via p38 MAPK and CD36. J Extracell Vesicles 2018, 7(1): 1432206.
- Hiemstra TF, Charles PD, Gracia T, Hester SS, Gatto L, Al-Lamki R, Floto RA, Su Y, Skepper JN, Lilley KS, et al: Human urinary exosomes as innate immune effectors. J Am Soc Nephrol 2014, 25(9): 2017-2027.
- Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin‐Smith GK, et al: Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018, 7(1): 1535750.
- Vaskova EA, Yang PC: Exosomes as natural nanocarriers for therapeutic and diagnostic use in cardiovascular diseases. Nanomed Ischemic Cardiom 2020: 71-88.
- Zhu S, Li S, Yi M, Li N, Wu K: Roles of microvesicles in tumor progression and clinical applications. Int J Nanomedicine 2021, 16: 7071-7090.
- Yuana Y, Levels J, Grootemaat A, Sturk A, Nieuwland R: Co-isolation of extracellular vesicles and high-density lipoproteins using density gradient ultracentrifugation. J Extracell Vesicles 2014, 3(1): 23262.
- Merchant ML, Rood IM, Deegens JKJ, Klein JB: Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery. Nat Rev Nephrol 2017, 13(12): 731-749.
- Park S, Lee K, Park IB, Kim NH, Cho S, Rhee WJ, Oh Y, Choi J, Nam S, Lee DH: The profiles of microRNAs from urinary extracellular vesicles (EVs) prepared by various isolation methods and their correlation with serum EV microRNAs. Diabetes Res Clin Pract 2020, 160: 108010.
- Gerlach JQ, Krüger A, Gallogly S, Hanley SA, Hogan MC, Ward CJ, Joshi L, Griffin MD: Surface Glycosylation Profiles of Urine Extracellular Vesicles. PLOS ONE 2013, 8(9): e74801.
- Serafini-Cessi F, Malagolini N, Cavallone D: Tamm-Horsfall glycoprotein: biology and clinical relevance. Am J Kidney Dis 2003, 42(4): 658-676.
- Jovine L, Qi H, Williams Z, Litscher E, Wassarman PM: The ZP domain is a conserved module for polymerization of extracellular proteins. Nat Cell Biol 2002, 4(6): 457-461.
- Alvarez ML, Khosroheidari M, Kanchi Ravi R, DiStefano JK: Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int 2012, 82(9): 1024-1032.
- Raj DAA, Fiume I, Capasso G, Pocsfalvi G: A multiplex quantitative proteomics strategy for protein biomarker studies in urinary exosomes. Kidney Int 2012, 81(12): 1263-1272.
- Chen CY, Hogan MC, Ward CJ: Chapter Thirteen - Purification of Exosome-Like Vesicles from Urine. In: Methods in Enzymology. Volume 524, edn. Edited by Marshall WF: Academic Press; 2013: 225-241.
- Rood IM, Deegens JKJ, Merchant ML, Tamboer WPM, Wilkey DW, Wetzels JFM, Klein JB: Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int 2010, 78(8): 810-816.
- Cheruvanky A, Zhou H, Pisitkun T, Kopp JB, Knepper MA, Yuen PS, Star RA: Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am J Physiol Renal Physiol 2007, 292(5): F1657-F1661.
- Merchant ML, Powell DW, Wilkey DW, Cummins TD, Deegens JK, Rood IM, McAfee KJ, Fleischer C, Klein E, Klein JB: Microfiltration isolation of human urinary exosomes for characterization by MS. Proteomics Clin Appl 2010, 4(1): 84-96.
- Gonzales PA, Zhou H, Pisitkun T, Wang NS, Star RA, Knepper MA, Yuen PS: Isolation and purification of exosomes in urine. Methods Mol Biol 2010, 641: 89-99.
- Barreiro K, Dwivedi OP, Leparc G, Rolser M, Delic D, Forsblom C, Groop PH, Groop L, Huber TB, Puhka M, et al: Comparison of urinary extracellular vesicle isolation methods for transcriptomic biomarker research in diabetic kidney disease. J Extracell Vesicles 2020, 10(2): e12038.
- Musante L, Tataruch D, Gu D, Benito-Martin A, Calzaferri G, Aherne S, Holthofer H: A simplified method to recover urinary vesicles for clinical applications and sample banking. Sci Rep 2014, 4(1): 7532.
- Ku A, Lim HC, Evander M, Lilja H, Laurell T, Scheding S, Ceder Y: Acoustic enrichment of extracellular vesicles from biological fluids. Anal Chem 2018, 90(13): 8011-8019.
- Hammarström B, Skov NR, Olofsson K, Bruus H, Wiklund M: Acoustic trapping based on surface displacement of resonance modes. J Acoust Soc Am 2021, 149(3): 1445-1453.
- Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P: Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 2014, 3: 26913.
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT: MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011, 13(4): 423-433.
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al: Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011, 108(12): 5003-5008.
- Rupert DLM, Claudio V, Lässer C, Bally M: Methods for the physical characterization and quantification of extracellular vesicles in biological samples. Biochim Biophys Acta Gen Subj 2017, 1861(1, Part A): 3164-3179.
- Cizmar P, Yuana Y: Detection and Characterization of Extracellular Vesicles by Transmission and Cryo-Transmission Electron Microscopy. In: Extracellular Vesicles: Methods and Protocols. https://doi.org/10.1007/978-1-4939-7253-1_18. Epub ahead of print., edn. Edited by Kuo WP, Jia S. New York, NY: Springer New York; 2017: 221-232.
- Yuana Y, Koning RI, Kuil ME, Rensen PCN, Koster AJ, Bertina RM, Osanto S: Cryo-electron microscopy of extracellular vesicles in fresh plasma. J Extracell Vesicles 2013, 2(1): 21494.
- Musante L, Bontha SV, La Salvia S, Fernandez-Piñeros A, Lannigan J, Le TH, Mas V, Erdbrügger U: Rigorous characterization of urinary extracellular vesicles (uEVs) in the low centrifugation pellet-a neglected source for uEVs. Sci Rep 2020, 10(1): 3701.
- Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJP, Hole P, Carr B, Redman CWG, Harris AL, Dobson PJ, et al: Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine 2011, 7(6): 780-788.
- McNicholas K, Li JY, Michael MZ, Gleadle JM: Albuminuria is not associated with elevated urinary vesicle concentration but can confound nanoparticle tracking analysis. Nephrology 2017, 22(11): 854-863.
- Hartjes TA, Mytnyk S, Jenster GW, van Steijn V, van Royen ME: Extracellular Vesicle Quantification and Characterization: Common Methods and Emerging Approaches. Bioengineering 2019, 6(1): 7.
- Colombo M, Raposo G, Théry C: Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu Rev Cell Dev Biol 2014, 30: 255-289.
- Ferlay J, Ervik M, Lam F, Colombet M, Mery L, Piñeros M, Znaor A, Soerjomataram I, Bray F: Global cancer observatory. Cancer today 2018, 23(7): 323-326.
- Qin Z, Xu Q, Hu H, Yu L, Zeng S: Extracellular vesicles in renal cell carcinoma: multifaceted roles and potential applications identified by experimental and computational methods. Front Oncol 2020, 10: 724.
- Raimondo F, Morosi L, Corbetta S, Chinello C, Brambilla P, Della Mina P, Villa A, Albo G, Battaglia C, Bosari S et al: Differential protein profiling of renal cell carcinoma urinary exosomes. Molecular BioSystems 2013, 9(6): 1220-1233.
- Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR, et al: Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 2014, 46(3): 225-233.
- Dudani S, De Velasco G, Wells JC, Gan CL, Donskov F, Porta C, Fraccon A, Pasini F, Lee JL, Hansen A, et al: Evaluation of clear cell, papillary, and chromophobe renal cell carcinoma metastasis sites and association with survival. JAMA network open 2021, 4(1): e2021869-e2021869.
- Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, Heng DY, Larkin J, Ficarra V: Renal cell carcinoma. Nat Rev Dis Primers 2017, 3(1): 1-19.
- Umberto C, Montorsi F: Renal cancer. Lancet 2016, 387(10021): 894-906.
- Bisbee CA, Zhang J, Owens J, Hussain S: Cryoablation for the Treatment of Kidney Cancer: Comparison With Other Treatment Modalities and Review of Current Treatment. Cureus 2022, 14(11): e31195.
- Rich BJ, Noy MA, Dal Pra A: Stereotactic body radiotherapy for localized kidney cancer. Curr Urol Rep 2022, 23(12): 371-381.
- Himbert D, Zeuschner P, Ayoubian H, Heinzelmann J, Stöckle M, Junker K: Characterization of CD147, CA9, and CD70 as tumor-specific markers on extracellular vesicles in clear cell renal cell carcinoma. Diagnostics 2020, 10(12): 1034.
- Yuan Z, Kolluri KK, Gowers KH, Janes SM: TRAIL delivery by MSC-derived extracellular vesicles is an effective anticancer therapy. J Extracell Vesicles 2017, 6(1): 1265291.
- Rezaeian A, Khatami F, Heidari Keshel S, Akbari MR, Mirzaei A, Gholami K, Mohammadi Farsani R, Aghamir SMK: The effect of mesenchymal stem cells-derived exosomes on the prostate, bladder, and renal cancer cell lines. Sci Rep 2022, 12(1): 20924.
- Valera VA, Merino MJ: Misdiagnosis of clear cell renal cell carcinoma. Nat Rev Urol 2011, 8(6): 321-333.
- Kooijmans SA, Schiffelers RM, Zarovni N, Vago R: Modulation of tissue tropism and biological activity of exosomes and other extracellular vesicles: New nanotools for cancer treatment. Pharmacol Res 2016, 111: 487-500.
- Zieren RC, Dong L, Clark DJ, Kuczler MD, Horie K, Moreno LF, Lih T-SM, Schnaubelt M, Vermeulen L, Zhang H: Defining candidate mRNA and protein EV biomarkers to discriminate ccRCC and pRCC from non-malignant renal cells in vitro. Med Oncol 2021, 38(9): 1-13.
- Grange C, Tapparo M, Collino F, Vitillo L, Damasco C, Deregibus MC, Tetta C, Bussolati B, Camussi G: Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res 2011, 71(15): 5346-5356.
- Zhang W, Ni M, Su Y, Wang H, Zhu S, Zhao A, Li G: MicroRNAs in serum exosomes as potential biomarkers in clear-cell renal cell carcinoma. Eur Urol Focus 2018, 4(3): 412-419.
- Wang X, Wang T, Chen C, Wu Z, Bai P, Li S, Chen B, Liu R, Zhang K, Li W, et al: Serum exosomal miR-210 as a potential biomarker for clear cell renal cell carcinoma. J Cell Biochem 2019, 120(2): 1492-1502.
- Fujii N, Hirata H, Ueno K, Mori J, Oka S, Shimizu K, Kawai Y, Inoue R, Yamamoto Y, Matsumoto H: Extracellular miR-224 as a prognostic marker for clear cell renal cell carcinoma. Oncotarget 2017, 8(66): 109877.
- Zhang Z, Hu J, Ishihara M, Sharrow AC, Flora K, He Y, Wu L: The miRNA-21-5p payload in exosomes from M2 macrophages drives tumor cell aggression via PTEN/Akt signaling in renal cell carcinoma. Int J Mol Sci 2022, 23(6): 3005.
- Horie K, Kawakami K, Fujita Y, Matsuda Y, Arai T, Suzui N, Miyazaki T, Koie T, Mizutani K, Ito M: Serum exosomal gamma-glutamyltransferase activity increased in patients with renal cell carcinoma with advanced clinicopathological features. Oncology 2020, 98(10): 734-742.
- Yoshino H, Tatarano S, Tamai M, Tsuruda M, Iizasa S, Arima J, Kawakami I, Fukumoto W, Kawahara I, Li G, et al: Exosomal microRNA-1 and MYO15A as a target for therapy and diagnosis in renal cell carcinoma. Biochem Biophys Res Commun 2022, 630: 71-76.
- Cech TR, Steitz JA: The noncoding RNA revolution—trashing old rules to forge new ones. Cell 2014, 157(1): 77-94.
- Qi P, Zhou X-y, Du X: Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer 2016, 15(1): 39.
- Pan Y, Lu X, Shu G, Cen J, Lu J, Zhou M, Huang K, Dong J, Li J, Lin H, et al: Extracellular vesicle-mediated transfer of LncRNA IGFL2-AS1 confers sunitinib resistance in renal cell carcinoma. Cancer Res 2023, 83(1): 103-116.
- Qu L, Ding J, Chen C, Wu Z-J, Liu B, Gao Y, Chen W, Liu F, Sun W, Li X-F, et al: Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer cell 2016, 29(5): 653-668.
- Erdbrügger U, Blijdorp CJ, Bijnsdorp IV, Borràs FE, Burger D, Bussolati B, Byrd JB, Clayton A, Dear JW, Falcón‐Pérez JM, et al: Urinary extracellular vesicles: A position paper by the Urine Task Force of the International Society for Extracellular Vesicles. J Extracell Vesicles 2021, 10(7): e12093.
- Boussios S, Devo P, Goodall IC, Sirlantzis K, Ghose A, Shinde SD, Papadopoulos V, Sanchez E, Rassy E, Ovsepian SV: Exosomes in the diagnosis and treatment of renal cell cancer. Int J Mol Sci 2023, 24(18): 14356.
- Blijdorp CJ, Tutakhel OA, Hartjes TA, van den Bosch TP, van Heugten MH, Rigalli JP, Willemsen R, Musterd-Bhaggoe UM, Barros ER, Carles-Fontana R, et al: Comparing approaches to normalize, quantify, and characterize urinary extracellular vesicles. J Am Soc Nephrol 2021, 32(5): 1210-1226.
- Blijdorp CJ, Hartjes TA, Wei KY, van Heugten MH, Bovée DM, Budde RP, van de Wetering J, Hoenderop JG, van Royen ME, Zietse R, et al: Nephron mass determines the excretion rate of urinary extracellular vesicles. J Extracell Vesicles 2022, 11(1): e12181.
- Raimondo F, Morosi L, Corbetta S, Chinello C, Brambilla P, Della Mina P, Villa A, Albo G, Battaglia C, Bosari S: Differential protein profiling of renal cell carcinoma urinary exosomes. Mol Biosyst 2013, 9(6): 1220-1233.
- US National Library of Medicine. ClinicalTrials.gov. 2021. Epub ahead of print. https://clinicaltrials.gov/ct2/show/NCT04053855.
- Barreiro K, Lay AC, Leparc G, Tran VDT, Rosler M, Dayalan L, Burdet F, Ibberson M, Coward RJ, Huber TB, et al: An in vitro approach to understand contribution of kidney cells to human urinary extracellular vesicles. J Extracell Vesicles 2023, 12(2): 12304.
- Kurahashi R, Kadomatsu T, Baba M, Hara C, Itoh H, Miyata K, Endo M, Morinaga J, Terada K, Araki K et al: MicroRNA-204-5p: A novel candidate urinary biomarker of Xp11.2 translocation renal cell carcinoma. Cancer Sci 2019, 110(6): 1897-1908.
- Huang T, Wang X, Yang X, Ji J, Wang Q, Yue X, Dong Z: Long non-coding RNA DUXAP8 enhances renal cell carcinoma progression via downregulating miR-126. Med Sci Monit 2018, 24: 7340.
- Kuczler MD, Zieren RC, Dong L, de Reijke TM, Pienta KJ, Amend SR: Advancements in the identification of EV derived mRNA biomarkers for liquid biopsy of clear cell renal cell carcinomas. Urology 2022, 160: 87-93.
- Marek-Bukowiec K, Konieczny A, Ratajczyk K, Czapor-Irzabek H, Górniak A, Kowal P: mRNA fingerprint of early-stage clear cell renal cell carcinoma identified in urinary exosomes by mRNA sequencing. Pol Arch Intern Med 2021, 131(6): 582-585.
- Qin Z, Hu H, Sun W, Chen L, Jin S, Xu Q, Liu Y, Yu L, Zeng S: miR-224-5p contained in urinary extracellular vesicles regulates PD-L1 expression by inhibiting cyclin D1 in renal cell carcinoma cells. Cancers 2021, 13(4): 618.
- Del Boccio P, Raimondo F, Pieragostino D, Morosi L, Cozzi G, Sacchetta P, Magni F, Pitto M, Urbani A: A hyphenated microLC‐Q‐TOF‐MS platform for exosomal lipidomics investigations: application to RCC urinary exosomes. Electrophoresis 2012, 33(4): 689-696.
Annals of urologic oncology
p-ISSN: 2617-7765, e-ISSN: 2617-7773
 Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript