Review Article | Open Access
A Potential Role of MicroRNA in the Renal Cancer and Its Tumor Microenvironment
Daniel Chikere Ali1, Siva Bharath Merugu2
1Maduka University, School of Pharmacy, Along Nsukka - Enugu New Road, Ekwegbe, Enugu State, Nigeria.
2Department of Oncology, Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC, USA.
Correspondence: Siva Bharath Merugu (Department of Oncology, Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC, USA; Email: bharath.niser@gmail.com).
Annals of Urologic Oncology 2025, 8(1): 30-39. https://doi.org/10.32948/auo.2025.01.25
Received: 18 Dec 2024 | Accepted: 21 Jan 2025 | Published online: 29 Jan 2025
Key words microRNAs, renal cell carcinoma, biomarkers, tumor microenvironment
A complex biological process involving gene mutations, genomic instability, and epigenetic alterations leads to the development of ccRCC [6]. The von Hippel-Lindau (VHL) gene is one of the important genes linked to the pathophysiology of ccRCC and is commonly mutated in this cancer type. VHL mutations cause hypoxia-inducible factors (HIFs) to become active, which changes the expression of angiogenic factors such platelet-derived growth factor (PDGF) and vascular endothelial growth factor (VEGF) [7]. The mammalian target of rapamycin (mTOR) signaling pathway is also triggered by VHL mutations, which increases HIF activity and encourages angiogenesis, ultimately propelling the development of ccRCC. These discoveries have aided in the creation of medications that target the mTOR pathway, PDGF/PDGF receptor (PDGFR), and VEGF/VEGF receptor (VEGFR) [8].
Despite these advances, reliable diagnostic and prognostic biomarkers for ccRCC have yet to be established for clinical use. Recently, the role of epigenetics in ccRCC, particularly non-coding RNAs (ncRNAs), has garnered significant attention. Many ncRNAs have been found to exhibit aberrant expression in various tumors, highlighting their critical role in tumor initiation and progression [9]. Exploring the functions of ncRNAs further may pave the way for improved early diagnosis and treatment strategies for ccRCC. In addition to examining how miRNA axes regulate the TME and highlighting their potential as novel biomarkers and therapeutic targets for RCC, this study attempts to give a summary of current developments regarding the involvement of the miRNA axis in RCC pathogenesis.
The fact that a single gene is usually regulated by numerous miRNAs and that each miRNA can target multiple genes with sequences complementary to its seed region further emphasizes the crucial role that miRNAs play in gene regulation [12]. An estimated one-third of the human genome is thought to be regulated by miRNAs, which also affect almost all important cellular functions [13]. The involvement of miRNAs in cancer pathogenesis has been extensively studied [14]. These molecules have been shown to impact essential aspects of cancer biology, including sustained cell proliferation, evasion of growth-inhibitory signals, and promotion of angiogenesis [15].
When miR-15a and miR-16-1 were discovered in a commonly deleted region in B-cell chronic lymphocytic leukemia, the significance of miRNAs in cancer was first brought to light [16]. Later studies found more genetic changes in miRNA-coding genes in a number of malignancies, such as breast cancer [17], ovarian cancer, lung cancer [18], and melanoma. Additionally, it has been demonstrated that oncogenes like c-Myc decrease tumor-suppressor miRNAs while modulating the expression of oncogenic miRNAs [19]. To learn more about the function miRNA expression profiles play in the pathophysiology of RCC, a number of studies have determined these profiles in a variety of biological samples [20].
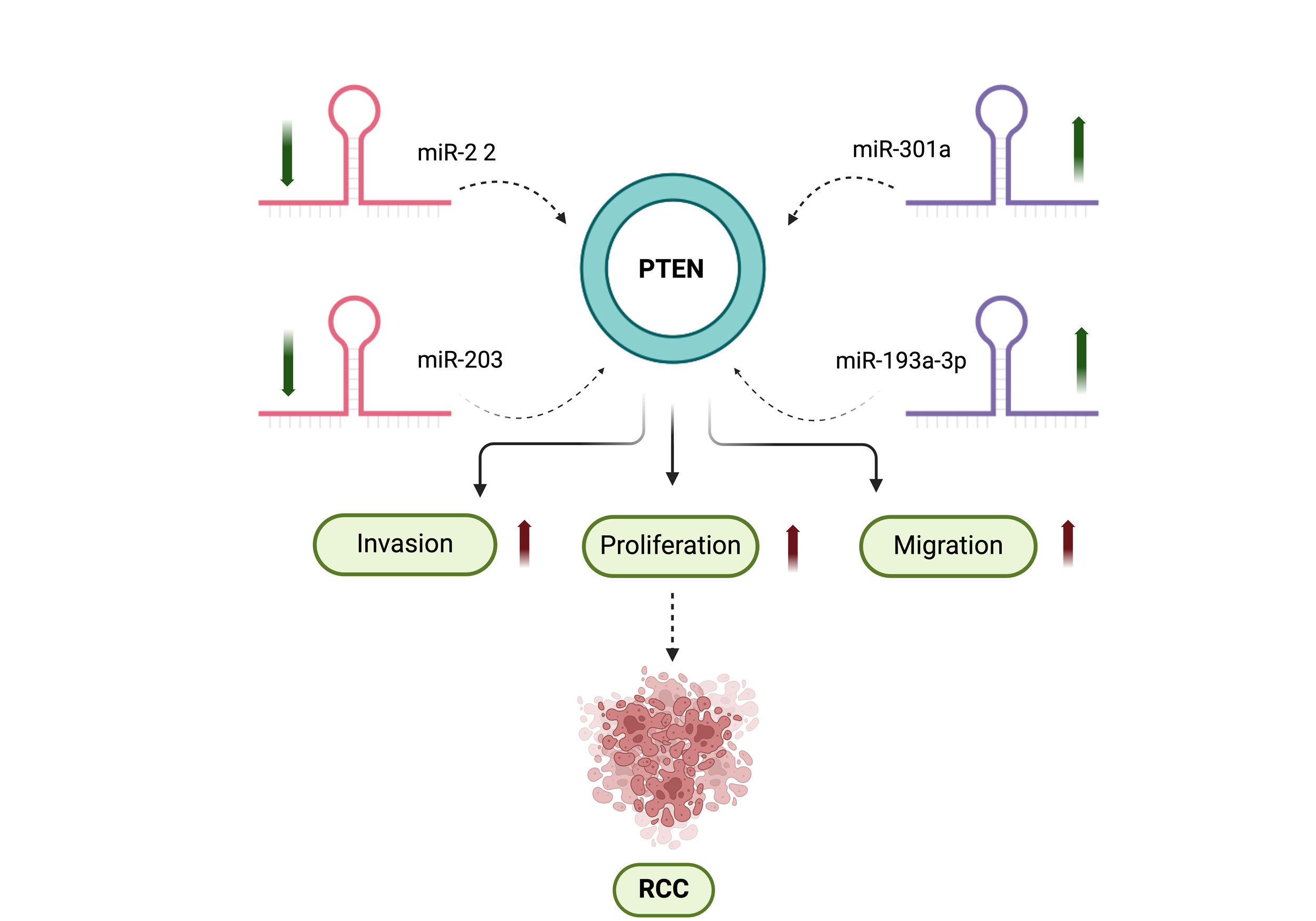 Figure 1. The scheme shows the interaction between miRNAs and the tumor-suppressor gene PTEN plays a crucial role in RCC. In RCC, miR-22 and miR-203 exhibit decreased expression, whereas miR-301 and miR-193a-3p are upregulated. These alterations in miRNA expression lead to the downregulation of PTEN, which in turn promotes increased cell proliferation, invasion, and migration.
Figure 1. The scheme shows the interaction between miRNAs and the tumor-suppressor gene PTEN plays a crucial role in RCC. In RCC, miR-22 and miR-203 exhibit decreased expression, whereas miR-301 and miR-193a-3p are upregulated. These alterations in miRNA expression lead to the downregulation of PTEN, which in turn promotes increased cell proliferation, invasion, and migration.
Another notable oncomiR, miR-301a, is upregulated in RCC cell lines and clinical samples. Its overexpression has been linked to advanced tumor stages and poor patient survival. Mechanistically, miR-301a directly targets the PTEN tumor suppressor [24]. Similarly, miR-22 and miR-193a-3p have also been shown to suppress PTEN expression in RCC cells [25]. Additionally, miR-1293 is upregulated in RCC cells, promoting their viability, migration, and invasiveness by inhibiting Hydrocyanic Oxidase 2 [26]. Table 1 summarizes the roles of upregulated miRNAs in RCC.
|
Table 1. miRNAs in RCC that are up-regulated (ANTTs: adjacent non-tumoral tissues). |
||||
|
miRNA |
Regulators |
Roles |
Samples |
Reference |
|
miR-21
|
TIMP3
|
Decreased miR-21 expression decreased cell invasion and migration and inhibited cells |
104 paired cancer tissues and ANTTs |
[88]
|
|
miR-210-3p
|
TWIST1
|
miR-210-3p promotes cell proliferation and tumorigenesis |
15 paired cancer tissues and ANTTs |
[89]
|
|
miR-29b
|
KIF1B
|
miR-29b increases cell proliferation and invasion, and suppresses apoptosis |
45 paired cancer tissues and ANTTs |
[90] |
|
miR-144-3p
|
ARID1A
|
miR-144-3p induces cell Proliferation and metastasis, in ccRCC by reducing ARID1A expression |
Tissues from 60 patients with ccRCC, 8 patients with nccRCC and 10 patients with renal hamartoma |
[91]
|
|
miR-106b-5p
|
SETD2
|
miR-106b-5p induces cells proliferation and inhibits apoptosis through reducing of SETD2 expression |
40 paired cancer tissues and ANTTs
|
[92] |
|
miR-122
|
Sprouty2
|
miR-122 induces cell proliferation by targeting Sprouty2 |
40 paired cancer tissues and ANTTs |
[93] |
|
miR-122
|
Occludin
|
miR-122 enhanced cell proliferation, migration and invasion |
90 paired cancer tissues and ANTTs
|
[94] |
|
miR-155
|
FOXO3a
|
miR-155 increased the proliferation, and nhibited apoptosis and cell cycle arrest |
20 paired cancer tissues and ANTTs
|
[95] |
|
miR-203a
|
GSK-3beta
|
miR-203a induces cell proliferation, migration, cell cycle, and suppresses apoptosis of RCC |
40 paired cancer tissues and ANTTs
|
[96] |
|
miR‐7
|
MEG3, RASL11B
|
miR‐7 induces progression of ccRCC |
72 paired samples from cancer tissues and ANTTs |
[97] |
|
miR-122
|
Dicer
|
miR‐122 induces EMT, migration and invasion in RCC |
148 cancer tissues and 60 ANTTs |
[98] |
|
miR-22
|
PTEN
|
Has a role in invasion |
480 paired ccRCC tissues and ANTTs and urine samples |
[99] |
|
miR-592
|
SPRY2
|
Has a role in proliferation, migration and invasion |
114 paired ccRCC tissues and ANTTs and urine samples |
[100] |
|
miR-671-5p
|
APC
|
Has a role in invasion and migration |
90 primary ccRCC tissues and 90 |
[101] |
|
miR-429
|
CRKL
|
Has a role in migration and invasion |
28 pairs of tumor and ANTTs |
[102] |
|
miR-301a
|
PTEN
|
miR-301a regulates PTEN expression |
516 tumor samples and 71 ANTTs |
[103] |
Numerous studies have shown that the Ras-Raf-MEK-ERK signaling pathway is essential to the initiation and spread of cancer [27]. For instance, RCC cells overexpress astrocyte-elevated gene-1 (AEG-1), a downstream gene of Ha-ras, which encourages cell invasion and proliferation. Interestingly, it has been demonstrated that miR-384 reverses these effects by specifically targeting AEG-1 [28]. According to another study, RCC has an increased level of p21-activated kinase 5 (PAK5), which reduces the metastasis-suppressive effects of miR-106a-5p [29]. Rho-associated protein kinase 1 (ROCK1) and Kirsten rat sarcoma viral oncogene (KRAS), are also highly expressed in RCC cells and aid in the growth of tumors. Accordingly, miR-199a and miR-532-5p can lessen these effects. Furthermore, it has been demonstrated that miR-532-5p suppresses P-ERK and ETS1 expression in vivo and inhibits tumor growth. In many types of cancer, ETS1 acts as an oncogene [30].
In RCC, miR-622/200b decreases phosphorylated ERK (P-ERK), a crucial element of the MAPK signaling pathway, while CCL18/LAMA4 increases it. MiR-622 can reverse the tumor progression caused by elevated CCL18 and LAMA4 expression in kidney carcinoma. Laminin subunit alpha-4 (LAMA4) and C-C motif chemokine 18 (CCL18) are both essential for the growth of tumors [31]. Furthermore, it has been observed that overexpression of miR-363 inhibits tumor growth in ccRCC [32]. Similarly, in RCC, spindle and kinetochore-associated protein 1 (SKA1) stimulates tumor growth, increases P-ERK1/2 and P-AKT levels. However, miR-10a-5p has the ability to reverse these effects. Numerous malignancies have been found to have SKA1 as an oncogene [33].
PI3K/AKT/mTOR signaling pathway
Renal cancer frequently exhibits dysregulation of the PI3K/Akt/mTOR signaling pathway [34]. This pathway's downstream target, Forkhead box protein M1 (FOXM1), is a member of the Forkhead box family and is highly expressed in RCC, which accelerates the growth of tumors. Notably, miR-149 and miR-320a can reverse these carcinogenic effects [35]. In RCC, oncogenes such as KIFC1, eIF4E, and HMGN5 upregulate the expression of PI3K and Akt, whereas miR-338-3p, miR-15a, and miR-488 can downregulate it. The advancement of renal carcinoma caused by these oncogenes is inhibited by overexpression of miR-338-3p, miR-15a, and miR-488 [36].
In RCC, SPOP can decrease PTEN expression, a crucial regulator of the PI3K/Akt pathway, whereas miR-520/372/373 and miR-203 can increase it. Through its targeting of SPOP, miR-520/372/373 overexpression inhibits the growth of tumors [37]. The tumor-suppressive effects of miR-148a are also reversed by AKT2, a member of the Akt family that is overexpressed in RCC. Via Rab14, a GTPase in the RAS oncogene family, miR-148a stimulates the production of TRAIL (tumor necrosis factor-related apoptosis-inducing ligand) and makes RCC cells more sensitive to cisplatin by lowering P-Akt and mTOR levels [38].
Additionally, splicing factor 2 (SF2) increases the expression of P-Akt and P-ERK in RCC, while miR-766-3p reduces their levels. Overexpression of miR-766-3p suppresses tumor growth by targeting SF2, a known promoter of carcinogenesis [39]. Another critical component of the Akt pathway, PDK1 is overexpressed in RCC, driving cell proliferation, but its effects are reversed by miR-375 [40]. Remarkably, miR-100 has been demonstrated to raise LC3 and LC3-II/LC3-I levels while downregulating mTOR and NOX4 expression. These modifications improve autophagy and lessen the aggression of RCC cells [41]. The connections between these compounds and their regulatory miRNAs are depicted in a Figure 2.
VEGF signaling pathway
Vascular endothelial growth factor (VEGF) is integral to tumor progression and angiogenesis [42]. In RCC, VEGFA is overexpressed and significantly promotes tumor growth and metastasis. However, its effects can be counteracted by miR-205-5p and miR-299-3p. miR-205-5p enhances RCC cells' sensitivity to treatments such as sunitinib, paclitaxel, 5-FU, and oxaliplatin by suppressing VEGFA expression [43]. Similarly, the growth hormone receptor (GHR) upregulates VEGF, thereby increasing the mobility of RCC cells, while this effect is reversed by miR-363. Additionally, GHR has been shown to positively correlate with RCC proliferation [44].
Another important miRNA, miR-218, has been shown to reduce tumor angiogenesis and decrease the migration of human umbilical vein endothelial cells (HUVECs) by suppressing VEGFA production and blocking the p-PI3K/p-Akt/p-mTOR signaling pathway. The downregulation of GRB2-associated binding protein 2 (GAB2), a crucial oncogene linked to several malignancies, mediates these effects [45]. Additionally, there is evidence that VEGF and fibroblast growth factor (FGF) play similar functions in the migration and activation of angioblasts during vascular development [46]. For example, FGF2 promotes invasion and tube formation in HUVECs within RCC. However, miR-148b-3p can counteract these pro-angiogenic effects by downregulating platelet-derived growth factor-BB/D (PDGF-BB/D), VEGFA, and FGF2 activity. In a number of malignancies, PDGF-BB and PDGF-D are recognized to act as strong pro-angiogenic mediators [47].
Metabolism/Immunity-related mechanism
Metabolic alterations within the TME contribute to immune evasion by generating immunosuppressive metabolites [48]. For example, overexpression of PLP2 in clear cell RCC (ccRCC) promotes tumor growth and mobility, enhances VEGFA expression, and facilitates lipid accumulation, an effect that can be counteracted by miR-765. PLP2, a newly identified gene upregulated in cancer, has been associated with oncogenic activity in breast cancer [49]. Studies have shown that RCC cells, like many other cancer forms, depend on aerobic glycolysis to produce ATP and have aberrant HIF stabilization [50]. In particular, ccRCC cells with high PDK1 expression increase the extracellular acidification rate (ECAR) in hypoxic environments, which boosts ATP synthesis and lowers the oxygen consumption rate (OCR) of tumor cells. MiR-409-3p can correct these metabolic alterations [51].
One of the key areas of research continues to be tumor immune evasion, which is a characteristic of cancer. MiR-497-5p can counteract the effect of overexpression of PD-L1, the ligand for the immunological checkpoint receptor PD-1, which promotes tumor growth in RCC [52]. Tumor formation is also influenced by TLRs, which control the innate immune feedback. For example, miR-216a targets TLR4 to inhibit RCC growth in vitro and in vivo [53].
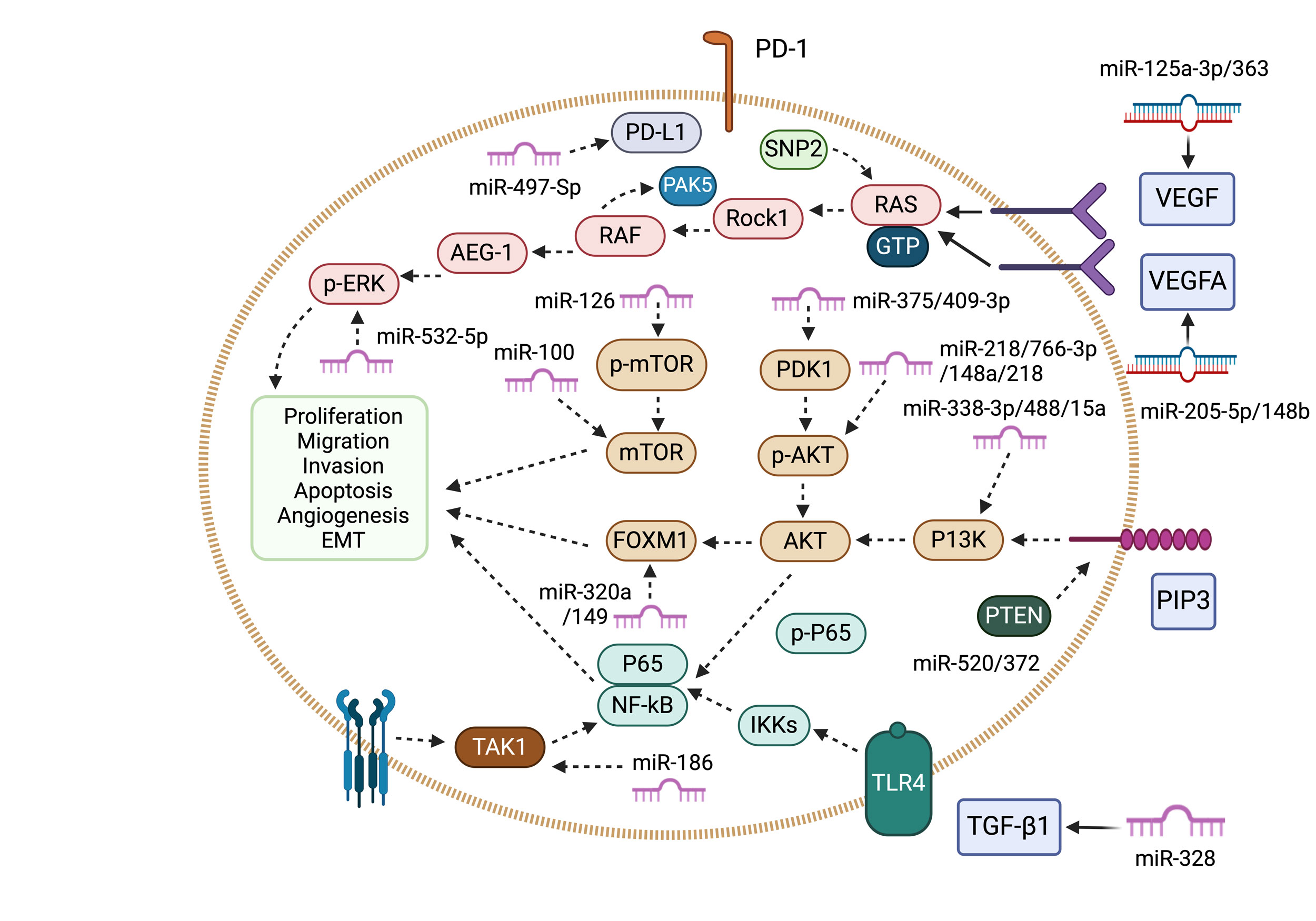 Figure 2. This diagram illustrates the role of tumor-suppressing miRNAs and their associated signaling pathways in renal cancer. miR-125a-3p/363 and miR-205-5p/148b-3p/299-3p/765 inhibit VEGF signaling by reducing VEGF and VEGFA expression. miR-199a, miR-106a-5p, miR-384, and others suppress the RAS/MAPK pathway by downregulating Rock1, PAK5, AEG-1, and p-ERK. Similarly, multiple miRNAs influence the PI3K/AKT/mTOR pathway by reducing PI3K, p-AKT, PDK1, and FOXM1 levels. miR-520/372/373/203 enhance PTEN expression, while miR-328 and miR-486-5p inhibit TGF-β signaling. Additionally, miR-186 and miR-216a regulate NF-κB signaling, and miR-497-5p targets PD-L1, affecting immune responses.
Figure 2. This diagram illustrates the role of tumor-suppressing miRNAs and their associated signaling pathways in renal cancer. miR-125a-3p/363 and miR-205-5p/148b-3p/299-3p/765 inhibit VEGF signaling by reducing VEGF and VEGFA expression. miR-199a, miR-106a-5p, miR-384, and others suppress the RAS/MAPK pathway by downregulating Rock1, PAK5, AEG-1, and p-ERK. Similarly, multiple miRNAs influence the PI3K/AKT/mTOR pathway by reducing PI3K, p-AKT, PDK1, and FOXM1 levels. miR-520/372/373/203 enhance PTEN expression, while miR-328 and miR-486-5p inhibit TGF-β signaling. Additionally, miR-186 and miR-216a regulate NF-κB signaling, and miR-497-5p targets PD-L1, affecting immune responses.
In many forms of cancer, the protein kinase mTOR is essential for controlling cell division and metabolism [54]. miR-92b-3p activates the mTOR signaling pathway and accelerates the development of ccRCC by decreasing the expression of TSC complex subunit 1 (TSC1) and increasing the phosphorylation of p70S6 kinase, a downstream target of TSC1. The role of TSC1 is to inhibit mTORC1 [55]. According to a different study, overexpression of miR-501-5p activates p-mTOR, which causes cell autophagy and p53 degradation in renal cancer, which aids in the tumor's growth in RCC [56]. Recent studies further reveal that IMPA2 (inositol monophosphatase 2) might reduce p-mTORC1 levels in ccRCC cells, establishing it as a possible biomarker for directing the use of mTOR inhibitors in treating metastatic ccRCC in clinical settings [57]. Furthermore, reduced expression of IMPA2 in ccRCC leads to decreased levels of N-cadherin and Slug, hindering tumor metastasis by downregulating miR-25-3p [58]. Altered cellular energetics is a characteristic of RCC and many other malignancies. For example, the pentose phosphate pathway (PPP) is typically increased in RCC cells, and G6PD, a major enzyme in this system, is a rate-limiting component. G6PD inhibition reduces the survival of RCC cells [59].
PI3K/AKT signaling pathway
One of the main characteristics of carcinogenesis in RCC is the high level of activation and frequent mutations in the PI3K/Akt signaling pathway [60]. In RCC, ST3GalIV downregulates p-PI3K and p-Akt expression, whereas miR-193a-3p and miR-224 upregulate them. Both miR-193a-3p and miR-224 target ST3GalIV (alpha-2,3-sialyltransferase IV), which aids in the growth of tumors. The production of α-2,3-sialic acid on cell surfaces, which is catalyzed by ST3GalIV, is intimately associated with the capacity for malignancies to spread [61]. Another study revealed that FRK (Fyn-related kinase) enhances p-PTEN levels, as FRK acts as a substrate for PTEN, although this effect is inhibited by miR-19 in ccRCC. Moreover, overexpression of miR-19 promotes cell proliferation in renal cancer by modulating FRK and PTEN [62]. FOXO3, a downstream target of the PI3K-Akt pathway is negatively regulated by miR-122 and highly expressed in ccRCC. In kidney cancer, miR-122 overexpression increases E-cadherin expression and stimulates tumor growth [63]. It's interesting to note that BTG3 (B-cell translocation gene 3) suppresses prostate cancer cells by lowering p-Akt levels [64]. By adversely controlling miR-142-5p, BTG3 inhibits cell growth in RCC [65] (Figure 3).
TGF-b/Wnt signaling pathway
Numerous developmental processes are regulated by TGF-β and Wnt signaling pathways, which are also linked to the development of some malignancies [66]. SMAD family member 4 (SMAD4), for example, is an essential part of the TGF-β pathway. In vitro, RCC tumor metastasis is inhibited by decreased SMAD4 expression; however, miR-452-5p can reverse this effect. Furthermore, miR-452-5p targets SMAD4 to decrease kidney cancer cells' susceptibility to tyrosine kinase inhibitors (TKIs) [67]. A member of the TGF-β superfamily, BMPR1B (bone morphogenetic protein receptor type 1B) has a role in the emergence of certain types of cancer [68]. Similarly, miR-1274a downregulates BMPR1B to increase ccRCC cell proliferation and decrease apoptosis [69]. Dickkopf1 (DKK1) and DKK3, both extracellular Wnt pathway inhibitors, have been demonstrated in earlier research to function as tumor suppressors in renal carcinoma [70]. Similarly, in RCC, miR-543 and miR-125b promote tumor growth by inhibiting DKK1 and DKK3, respectively. Additionally, renal cancer cells are less sensitive to chemotherapeutic medications like doxorubicin and sunitinib when miR-125b is overexpressed [71] (Figure 3).
NF-kB signaling pathway
In renal cancer, miR-146b-5p lowers the protein levels of TRAF6 (TNF receptor-associated factor 6) and NF-κB (p65). Additionally, miR-146b-5p enhances tumor growth by reducing serum IFN-γ levels and altering TRAF6. In the NF-κB pathway, TRAF6 functions as a signal transducer, and IFN-γ has been utilized to treat ovarian cancer [72]. It's interesting to note that KLF6 has been demonstrated to suppress the growth of glioblastoma by reducing the localization of p65 [73]. Similarly, in ccRCC, overexpression of KLF6 reduces tumor development and raises p21 levels. However, p21 has been demonstrated to deactivate the NF-κB pathway in prostate cancer, and miR-543 can restore this impact [74]. According to earlier research, TNF deactivates the NF-κB pathway, which results in both necroptosis and apoptosis [75]. In contrast, miR-381-3p has no discernible effect on TNF-induced NF-κB activation but suppresses TNF-induced apoptosis and necroptosis in renal cancer. This encourages tumor growth and points to a dismal prognosis for pRCC patients. These results suggest that different cell types may have distinct functions for the NF-κB pathway. A key regulatory protein in planned cell necroptosis is RIPK3 [76] (Figure 3).
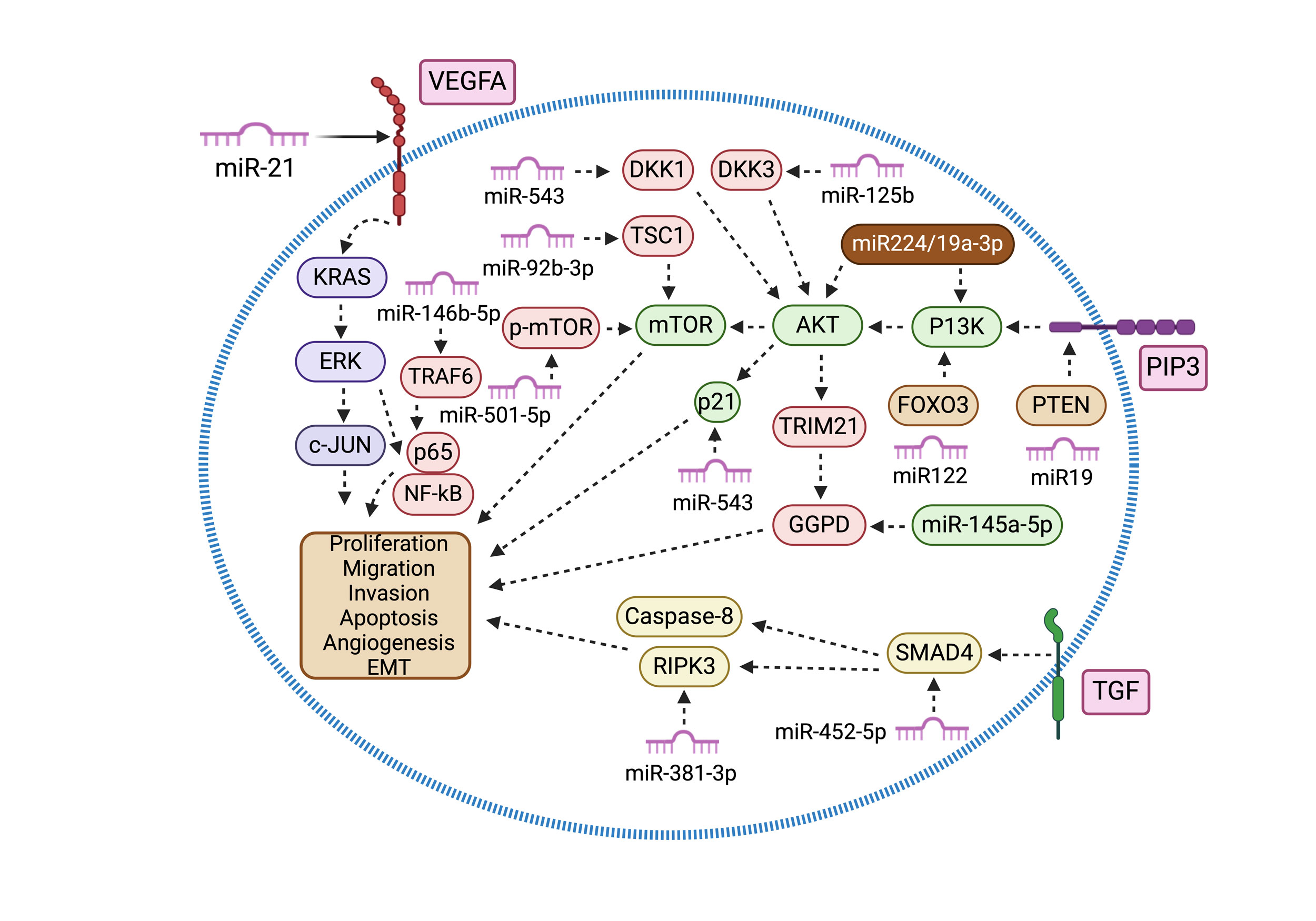 Figure 3. This diagram highlights oncogenic miRNAs and their involvement in signaling pathways in renal cancer. miR-224/193a-3p and miR-501-5p enhance PI3K/AKT/mTOR signaling by upregulating PI3K, AKT, and p-mTOR. Conversely, miR-19, miR-122, and miR-92b-3p suppress PTEN, FOXO3, and TSC1. miR-21 and miR-223-3p promote VEGF and RAS/MAPK pathways by increasing VEGFA/c-jun and KRAS levels. Additionally, miR-125b and miR-543 activate Wnt signaling by downregulating DKK3 and DKK1, while miR-452-5p enhances TGF-β signaling by reducing SMAD4 expression. miR-146a-5p and miR-146b-5p also play roles in PPP metabolism and inflammation by targeting G6PD and TRAF6.
Figure 3. This diagram highlights oncogenic miRNAs and their involvement in signaling pathways in renal cancer. miR-224/193a-3p and miR-501-5p enhance PI3K/AKT/mTOR signaling by upregulating PI3K, AKT, and p-mTOR. Conversely, miR-19, miR-122, and miR-92b-3p suppress PTEN, FOXO3, and TSC1. miR-21 and miR-223-3p promote VEGF and RAS/MAPK pathways by increasing VEGFA/c-jun and KRAS levels. Additionally, miR-125b and miR-543 activate Wnt signaling by downregulating DKK3 and DKK1, while miR-452-5p enhances TGF-β signaling by reducing SMAD4 expression. miR-146a-5p and miR-146b-5p also play roles in PPP metabolism and inflammation by targeting G6PD and TRAF6.
Extracellular miRNAs, including those in serum or urine, have become interesting biomarkers for RCC diagnosis and prognosis since they are likewise dysregulated in RCC patients. For example, it was possible to distinguish RCC from healthy controls by observing that serum miR-378 levels were higher and miR-451 levels were lower in RCC patients. With an AUC of 0.86 and a sensitivity of 81% and specificity of 83%, the combination of these two miRNAs enhanced stratification [82]. Patients with RCC had elevated serum levels of miR-1233, which demonstrated a sensitivity of 77.4% and specificity of 37.6%. One possible biomarker for RCC has been shown to be miR-1233 [83]. Furthermore, blood levels of miR-210 were considerably greater in ccRCC patients than in healthy controls, indicating that miR-210 was elevated in the early stages of the disease. For diagnostic purposes, this miRNA showed 81.0% sensitivity and 79.4% specificity [84, 85]. While miR-15a was nearly undetectable in oncocytoma, other tumors, and urinary tract inflammation, it was found to be elevated in RCC patients' urine samples [86]. Given their excellent diagnostic accuracy in RCC patients, miRNAs may be used as next-generation disease detection biomarkers. To confirm these findings and establish their regular clinical use, however, extensive research and additional developments are required [87].
None.
Ethical policy
Non applicable.
Availability of data and materials
All data generated or analysed during this study are included in this publication.
Author contributions
DCA searched academic literature, wrote the draft manuscript and draw the figures; SBM supervised the review writting progress and approved the final manuscript submission.
Competing interests
None.
Funding
None.
- Siegel RL, Miller KD, Jemal A: Cancer statistics, 2018. CA Cancer J Clin 2018, 72(1): 7-33.
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F: Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021, 71(3): 209-249.
- Bahadoram S, Davoodi M, Hassanzadeh S, Bahadoram M, Barahman M, Mafakher L: Renal cell carcinoma: an overview of the epidemiology, diagnosis, and treatment. G Ital Nefrol 2022, 39(3): 2022-vol3.
- Liu X, Hao Y, Yu W, Yang X, Luo X, Zhao J, Li J, Hu X, Li L: Long non-coding RNA emergence during renal cell carcinoma tumorigenesis. Cell Physiol Biochem 2018, 47(2): 735-746.
- Gilbert NJ: Surgical treatment of pulmonary metastases in metastatic renal cell carcinoma. Aktuelle Urol 2020, 51(3): 271-274.
- Weaver C, Bin Satter K, Richardson KP, Tran LK, Tran PM, Purohit S: Diagnostic and prognostic biomarkers in renal clear cell carcinoma. Biomedicines 2022, 10(11): 2953.
- Lai Y, Zhao Z, Zeng T, Liang X, Chen D, Duan X, Zeng G, Wu W: Crosstalk between VEGFR and other receptor tyrosine kinases for TKI therapy of metastatic renal cell carcinoma. Cancer cell Int 2018, 18: 31.
- Dell’Atti L, Bianchi N, Aguiari G: New therapeutic interventions for kidney carcinoma: looking to the future. Cancers 2022, 14(15): 3616.
- Kulkarni P, Dasgupta P, Hashimoto Y, Shiina M, Shahryari V, Tabatabai ZL, Yamamura S, Tanaka Y, Saini S, Dahiya R et al: A lncRNA TCL6-miR-155 interaction regulates the Src-Akt-EMT network to mediate kidney cancer progression and metastasis. Cancer Res 2021, 81(6): 1500-1512.
- Bracken CP, Scott HS, Goodall GJ: A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet 2016, 17(12): 719-732.
- Ha M, Kim VN: Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014, 15(8): 509-524.
- Kehl T, Backes C, Kern F, Fehlmann T, Ludwig N, Meese E, Lenhof HP, Keller A: About miRNAs, miRNA seeds, target genes and target pathways. Oncotarget 2017, 8(63): 107167-107175.
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP: The impact of microRNAs on protein output. Nature 2008, 455(7209): 64-71.
- Jansson MD, Lund AH. MicroRNA and cancer: Mol Oncol 2012, 6(6): 590-610.
- Peng Y, Croce CM: The role of MicroRNAs in human cancer. Signal Transduct Target Ther 2016, 1: 15004.
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K et al: Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci 2002, 99(24): 15524-15529.
- Calin GA, Croce CM: MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene 2006, 25(46): 6202-6210.
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR et al: microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci 2006, 103(24): 9136-9141.
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT: Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 2008, 40(1): 43-50.
- Mytsyk Y, Dosenko V, Skrzypczyk MA, Borys Y, Diychuk Y, Kucher A, Kowalskyy V, Pasichnyk S, Mytsyk O, Manyuk L et al: Potential clinical applications of microRNAs as biomarkers for renal cell carcinoma. Cent European J Urol 2018, 71(3): 295-303.
- Li J, Huang JH, Qu QH, Xia Q, Wang DS, Jin L, Sheng C: Evaluating the microRNA-target gene regulatory network in renal cell carcinomas, identification for potential biomarkers and critical pathways. Int J Clin Exp Med 2015, 8(5): 7209-7219.
- Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F et al: Micro-RNA profiling in kidney and bladder cancers. InUrologic Oncology: Seminars and Original Investigations 2007, 25(5): 387-392.
- Wulfken LM, Moritz R, Ohlmann C, Holdenrieder S, Jung V, Becker F, Herrmann E, Walgenbach-Bruenagel G, von Ruecker A, Mueller SC et al: MicroRNAs in renal cell carcinoma: diagnostic implications of serum miR-1233 levels. PloS One 2011, 6(9): e25787.
- Li J, Jiang D, Zhang Q, Peng S, Liao G, Yang X, Tang J, Xiong H, Pang J: MiR-301a promotes cell proliferation by repressing PTEN in renal cell carcinoma. Cancer Manag Res 2020, 12: 4309-4320.
- Gong X, Zhao H, Saar M, Peehl DM, Brooks JD: miR-22 regulates invasion, gene expression and predicts overall survival in patients with clear cell renal cell carcinoma. Kidney Cancer 2019, 3(2): 119-132.
- Liu XL, Pan WG, Li KL, Mao YJ, Liu SD, Zhang RM: miR-1293 suppresses tumor malignancy by targeting hydrocyanic oxidase 2: therapeutic potential of a miR-1293/hydrocyanic oxidase 2 axis in renal cell carcinoma. Cancer Biother Radiopharm 2020, 35(5): 377-386.
- Kim EK, Choi EJ: Compromised MAPK signaling in human diseases: an update. Arch Toxicol 2015, 89(6): 867-882.
- Song H, Rao Y, Zhang G, Kong X: MicroRNA-384 inhibits the growth and invasion of renal cell carcinoma cells by targeting astrocyte elevated gene 1. Oncol Res 2018, 26(3): 457-466.
- Pan YJ, Wei LL, Wu XJ, Huo FC, Mou J, Pei DS: MiR-106a-5p inhibits the cell migration and invasion of renal cell carcinoma through targeting PAK5. Cell Death Dis 2017, 8(10): e3155.
- Zhai W, Ma J, Zhu R, Xu C, Zhang J, Chen Y, Chen Z, Gong D, Zheng J, Chen C et al: MiR-532-5p suppresses renal cancer cell proliferation by disrupting the ETS1-mediated positive feedback loop with the KRAS-NAP1L1/P-ERK axis. Br J Cancer 2018, 119(5): 591-604.
- Li Y, Guan B, Liu J, Zhang Z, He S, Zhan Y, Su B, Han H, Zhang X, Wang B et al: MicroRNA-200b is downregulated and suppresses metastasis by targeting LAMA4 in renal cell carcinoma. EBioMedicine 2019, 44: 439-451.
- Xie Y, Chen L, Gao Y, Ma X, He W, Zhang Y, Zhang F, Fan Y, Gu L, Li P et al: miR-363 suppresses the proliferation, migration and invasion of clear cell renal cell carcinoma by downregulating S1PR1. Cancer Cell Int 2020, 20: 227.
- Arai T, Okato A, Kojima S, Idichi T, Koshizuka K, Kurozumi A, Kato M, Yamazaki K, Ishida Y, Naya Y et al: Regulation of spindle and kinetochore‐associated protein 1 by antitumor miR‐10a‐5p in renal cell carcinoma. Cancer Sci 2017, 108(10): 2088-2101.
- Pal SK, Quinn DI: Differentiating mTOR inhibitors in renal cell carcinoma. Cancer Treat Rev 2013, 39(7): 709-719.
- Zhao S, Wang Y, Lou Y, Wang Y, Sun J, Luo M, Li W, Miao L: MicroRNA320a suppresses tumour cell proliferation and invasion of renal cancer cells by targeting FoxM1. Oncol Rep 2018, 40(4): 1917-1926.
- Wei X, Yu L, Kong X: miR-488 inhibits cell growth and metastasis in renal cell carcinoma by targeting HMGN5. OncoTargets Ther 2018, 11: 2205-2216.
- Ding M, Lu X, Wang C, Zhao Q, Ge J, Xia Q, Wang J, Zen K, Zhang CY, Zhang C et al: The E2F1–miR-520/372/373–SPOP Axis Modulates Progression of Renal Carcinoma. Cancer Res 2018, 78(24): 6771-6784.
- Kim EA, Kim TG, Sung EG, Song IH, Kim JY, Doh KO, Lee TJ: miR-148a increases the sensitivity to cisplatin by targeting Rab14 in renal cancer cells. Int J Oncol 2017, 50(3): 984-992.
- Chen C, Xue S, Zhang J, Chen W, Gong D, Zheng J, Ma J, Xue W, Chen Y, Zhai W et al: DNA‐methylation‐mediated repression of miR‐766‐3p promotes cell proliferation via targeting SF2 expression in renal cell carcinoma. Int J Cancer 2017, 141(9): 1867-1878.
- Wang J, Sun X: MicroRNA-375 inhibits the proliferation, migration and invasion of kidney cancer cells by triggering apoptosis and modulation of PDK1 expression. EnvironToxicol Pharmacol 2018, 62: 227-233.
- Liu X, Zhong L, Li P, Zhao P: MicroRNA‐100 enhances autophagy and suppresses migration and invasion of renal cell carcinoma cells via disruption of NOX4‐dependent mTOR pathway. Clin Transl Sci 2022, 15(2): 567-575.
- Goel HL, Mercurio AM: VEGF targets the tumour cell. Nat Rev Cancer 2013, 13(12): 871-882.
- Li Y, Zheng D, Pan L, Dai Y, Cai S, Zhao L, Zhu H: Knockdown of TUG1 by shRNA inhibited renal cell carcinoma formation by miR-299–3p/VEGF axis in vitro and in vivo. Eur J Pharmacol 2019, 860: 172536.
- Huang Y, Zhou J, Deng Y, Li G, He S, Li H, Liu L: MiR-363: A potential biomarker of kidney diseases. Clin Chim Acta 2024, 567: 120049.
- Mu L, Guan B, Tian J, Li X, Long Q, Wang M, Wang W, She J, Li X, Wu D et al: MicroRNA218 inhibits tumor angiogenesis of human renal cell carcinoma by targeting GAB2 Corrigendum in/10.3892/or. 2022.8406. Oncol Rep 2020, 44(5): 1961-1970.
- Poole TJ, Finkelstein EB, Cox CM: The role of FGF and VEGF in angioblast induction and migration during vascular development. Dev Dyn 2001, 220(1): 1-7.
- Zhang H, Ye Q, Du Z, Huang M, Zhang M, Tan H: MiR-148b-3p inhibits renal carcinoma cell growth and pro-angiogenic phenotype of endothelial cell potentially by modulating FGF2. Biomed Pharmacother 2018, 107: 359-367.
- Hanahan D, Weinberg RA: Hallmarks of cancer: the next generation. Cell 2011, 144(5): 646-674.
- Xiao W, Wang C, Chen K, Wang T, Xing J, Zhang X, Wang X: MiR-765 functions as a tumour suppressor and eliminates lipids in clear cell renal cell carcinoma by downregulating PLP2. EBioMedicine 2020, 51: 102622.
- Chan DA, Sutphin PD, Nguyen P, Turcotte S, Lai EW, Banh A, Reynolds GE, Chi JT, Wu J, Solow-Cordero DE et al: Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci Transl Med 2011, 3(94): 94ra70.
- Wang Y, He Y, Bai H, Dang Y, Gao J, Lv P: Phosphoinositide‐dependent kinase 1-associated glycolysis is regulated by miR‐409‐3p in clear cell renal cell carcinoma. J Cell Biochem 2019, 120(1): 126-134.
- Qu F, Ye J, Pan X, Wang J, Gan S, Chu C, Chu J, Zhang X, Liu M, He H et al: MicroRNA-497-5p down-regulation increases PD-L1 expression in clear cell renal cell carcinoma. J Drug Target 2019, 27(1): 67-74.
- Wang W, Zhao E, Yu Y, Geng B, Zhang W, Li X: MiR-216a exerts tumor-suppressing functions in renal cell carcinoma by targeting TLR4. Am J Cancer Res 2018, 8(3): 476-488.
- Hua H, Kong Q, Zhang H, Wang J, Luo T, Jiang Y: Targeting mTOR for cancer therapy. J Hematol Oncol 2019, 12(1): 71.
- Wang C, Uemura M, Tomiyama E, Matsushita M, Koh Y, Nakano K, Hayashi Y, Ishizuya Y, Jingushi K, Kato T et al: MicroRNA‐92b‐3p is a prognostic oncomiR that targets TSC1 in clear cell renal cell carcinoma. Cancer Sci 2020, 111(4): 1146-1155.
- Patergnani S, Guzzo S, Mangolini A, dell’Atti L, Pinton P, Aguiari G: The induction of AMPK-dependent autophagy leads to P53 degradation and affects cell growth and migration in kidney cancer cells. Exp Cell Res 2020, 395(1): 112190.
- Kuei CH, Lin HY, Lee HH, Lin CH, Zheng JQ, Chen KC, Lin YF: IMPA2 downregulation enhances mTORC1 activity and restrains autophagy initiation in metastatic clear cell renal cell carcinoma. J Clin Med 2020, 9(4): 956.
- Lin YF, Chou JL, Chang JS, Chiu IJ, Chiu HW, Lin YF: Dysregulation of the miR-25-IMPA2 axis promotes metastatic progression in clear cell renal cell carcinoma. EBioMedicine 2019, 45: 220-230.
- Bogusławska J, Popławski P, Alseekh S, Koblowska M, Iwanicka-Nowicka R, Rybicka B, Kędzierska H, Głuchowska K, Hanusek K, Tański Z et al: MicroRNA-mediated metabolic reprograming in renal cancer. Cancers 2019, 11(12): 1825.
- Guo H, German P, Bai S, Barnes S, Guo W, Qi X, Lou H, Liang J, Jonasch E, Mills GB et al: The PI3K/AKT pathway and renal cell carcinoma. J Genet Genomics 2015, 42(7): 343-353.
- Pan Y, Hu J, Ma J, Qi X, Zhou H, Miao X, Zheng W, Jia L: MiR‐193a‐3p and miR‐224 mediate renal cell carcinoma progression by targeting alpha‐2, 3‐sialyltransferase IV and the phosphatidylinositol 3 kinase/Akt pathway. Mol Carcinog 2018, 57(8): 1067-1077.
- Jing ZF, Bi JB, Li ZL, Liu XK, Li J, Zhu YY, Zhang XT, Zhang Z, Li ZH, Kong CZ et al: miR-19 promotes the proliferation of clear cell renal cell carcinoma by targeting the FRK-PTEN axis. OncoTargets Ther 2019, 12: 2713-2727.
- Nie W, Ni D, Ma X, Zhang Y, Gao Y, Peng C, Zhang X: miR122 promotes proliferation and invasion of clear cell renal cell carcinoma by suppressing Forkhead box O3 Corrigendum in/10.3892/ijo. 2019.4694. Int J Oncol 2019, 54(2): 559-571.
- Cheng YC, Chen PH, Chiang HY, Suen CS, Hwang MJ, Lin TY, Yang HC, Lin WC, Lai PL, Shieh SY et al: Candidate tumor suppressor B-cell translocation gene 3 impedes neoplastic progression by suppression of AKT. Cell Death Dis 2015, 6(1): e1584.
- Liu L, Liu S, Duan Q, Chen L, Wu T, Qian H, Yang S, Xin D, He Z, Guo Y et al: MicroRNA-142-5p promotes cell growth and migration in renal cell carcinoma by targeting BTG3. Am J Transl Res 2017, 9(5): 2394-2402.
- Attisano L, Labbé E: TGFβ and Wnt pathway cross-talk. Cancer Metastasis Rev 2004, 23(1-2): 53-61.
- Zhai W, Li S, Zhang J, Chen Y, Ma J, Kong W, Gong D, Zheng J, Xue W, Xu Y et al: Sunitinib-suppressed miR-452-5p facilitates renal cancer cell invasion and metastasis through modulating SMAD4/SMAD7 signals. Mol Cancer 2018, 17(1): 157.
- de Vinuesa AG, Abdelilah-Seyfried S, Knaus P, Zwijsen A, Bailly S: BMP signaling in vascular biology and dysfunction. Cytokine Growth Factor Rev 2016, 27: 65-79.
- Yoshino H, Yonezawa T, Yonemori M, Miyamoto K, Sakaguchi T, Sugita S, Osako Y, Tatarano S, Nakagawa M, Enokida H et al: Downregulation of microRNA-1274a induces cell apoptosis through regulation of BMPR1B in clear cell renal cell carcinoma. Oncol Rep 2018, 39(1): 173-181.
- Ueno K, Hirata H, Majid S, Chen Y, Zaman MS, Tabatabai ZL, Hinoda Y, Dahiya R: Wnt antagonist DICKKOPF‐3 (Dkk‐3) induces apoptosis in human renal cell carcinoma. Mol Carcinog 2011, 50(6): 449-457.
- Yan L, Ding B, Liu H, Zhang Y, Zeng J, Hu J, Yao W, Yu G, An R, Chen Z et al: Inhibition of SMYD2 suppresses tumor progression by down-regulating microRNA-125b and attenuates multi-drug resistance in renal cell carcinoma. Theranostics 2019, 9(26): 8377-8391.
- Meng G, Li G, Yang X, Xiao N: Inhibition of miR146b‐5p suppresses CT‐guided renal cell carcinoma by targeting TRAF6. J Cell Biochem 2019, 120(2): 2382-2390.
- Masilamani AP, Ferrarese R, Kling E, Thudi NK, Kim H, Scholtens DM, Dai F, Hadler M, Unterkircher T, Platania L et al: KLF6 depletion promotes NF-κB signaling in glioblastoma. Oncogene 2017, 36(25): 3562-3575.
- Lee NJ, Oh JH, Ban JO, Shim JH, Lee HP, Jung JK, Ahn BW, Yoon DY, Han SB, Ham YW et al: 4‐O‐methylhonokiol, a PPARγ agonist, inhibits prostate tumour growth: p21‐mediated suppression of NF‐κB activity. Br J Pharmacol 2013, 168(5): 1133-1145.
- Geng J, Ito Y, Shi L, Amin P, Chu J, Ouchida AT, Mookhtiar AK, Zhao H, Xu D, Shan B et al: Regulation of RIPK1 activation by TAK1-mediated phosphorylation dictates apoptosis and necroptosis. Nat Commun 2017, 8(1): 359.
- Zhao C, Zhou Y, Ran Q, Yao Y, Zhang H, Ju J, Yang T, Zhang W, Yu X, He S et al: MicroRNA-381-3p functions as a dual suppressor of apoptosis and necroptosis and promotes proliferation of renal cancer cells. Front Cell Dev Biol 2020, 8: 290.
- Powers MP, Alvarez K, Kim HJ, Monzon FA: Molecular classification of adult renal epithelial neoplasms using microRNA expression and virtual karyotyping. Diagn Mol Pathol 2011, 20(2): 63-70.
- Chen X, Wang X, Ruan A, Han W, Zhao Y, Lu X, Xiao P, Shi H, Wang R, Chen L et al: miR-141 is a key regulator of renal cell carcinoma proliferation and metastasis by controlling EphA2 expression. Clin Cancer Res 2014, 20(10): 2617-2630.
- Faragalla H, Youssef YM, Scorilas A, Khalil B, White NM, Mejia-Guerrero S, Khella H, Jewett MA, Evans A, Lichner Z et al: The clinical utility of miR-21 as a diagnostic and prognostic marker for renal cell carcinoma. J Mol Diagn 2012, 14(4): 385-392.
- Silva-Santos RM, Costa-Pinheiro P, Luis A, Antunes L, Lobo F, Oliveira J, Henrique R, Jerónimo C: MicroRNA profile: a promising ancillary tool for accurate renal cell tumour diagnosis. Br J Cancer 2013, 109(10): 2646-2653.
- Youssef YM, White NM, Grigull J, Krizova A, Samy C, Mejia-Guerrero S, Evans A, Yousef GM: Accurate molecular classification of kidney cancer subtypes using microRNA signature. Eur Urol 2011, 59(5): 721-730.
- Redova M, Poprach A, Nekvindova J, Iliev R, Radova L, Lakomy R, Svoboda M, Vyzula R, Slaby O: Circulating miR-378 and miR-451 in serum are potential biomarkers for renal cell carcinoma. J Transl Med 2012, 10: 55.
- Wulfken LM, Moritz R, Ohlmann C, Holdenrieder S, Jung V, Becker F, Herrmann E, Walgenbach-Bruenagel G, von Ruecker A, Mueller SC et al: MicroRNAs in renal cell carcinoma: diagnostic implications of serum miR-1233 levels. PloS One 2011, 6(9): e25787.
- Iwamoto H, Kanda Y, Sejima T, Osaki M, Okada F, Takenaka A: Serum miR-210 as a potential biomarker of early clear cell renal cell carcinoma. Int J Oncol 2013, 44(1): 53-58.
- Zhao A, Li G, Péoc'h M, Genin C, Gigante M: Serum miR-210 as a novel biomarker for molecular diagnosis of clear cell renal cell carcinoma. Exp Mol Pathol 2013, 94(1): 115-120.
- von Brandenstein M, Pandarakalam JJ, Kroon L, Loeser H, Herden J, Braun G, Wendland K, Dienes HP, Engelmann U, Fries JW et al: MicroRNA 15a, inversely correlated to PKCα, is a potential marker to differentiate between benign and malignant renal tumors in biopsy and urine samples. Am J Pathol 2012, 180(5): 1787-1797.
- Gao Y, Zhao H, Lu Y, Li H, Yan G: MicroRNAs as potential diagnostic biomarkers in renal cell carcinoma. Tumor Biol 2014, 35(11): 11041-11050.
- Chen J, Gu Y, Shen W: MicroRNA-21 functions as an oncogene and promotes cell proliferation and invasion via TIMP3 in renal cancer. Eur Rev Med Pharmacol Sci 2017, 21 (20): 4566-4576.
- Yoshino H, Yonemori M, Miyamoto K, Tatarano S, Kofuji S, Nohata N, Nakagawa M, Enokida H: microRNA-210-3p depletion by CRISPR/Cas9 promoted tumorigenesis through revival of TWIST1 in renal cell carcinoma. Oncotarget 20178(13): 20881-20894.
- Xu Y, Zhu J, Lei Z, Wan L, Zhu X, Ye F, Tong Y: Expression and functional role of miR-29b in renal cell carcinoma. Int J Clin Exp Pathol 2015, 8(11): 14161-14170.
- Xiao W, Lou N, Ruan H, Bao L, Xiong Z, Yuan C, Tong J, Xu G, Zhou Y, Qu Y et al: Mir-144-3p promotes cell proliferation, metastasis, sunitinib resistance in clear cell renal cell carcinoma by downregulating ARID1A. Cell Physiol Biochem 2017, 43(6): 2420-2433.
- Xiang W, He J, Huang C, Chen L, Tao D, Wu X, Wang M, Luo G, Xiao X, Zeng F et al: miR-106b-5p targets tumor suppressor gene SETD2 to inactive its function in clear cell renal cell carcinoma. Oncotarget 2015, 6(6): 4066-4079.
- Wang Z, Qin C, Zhang J, Han Z, Tao J, Cao Q, Zhou W, Xu Z, Zhao C, Tan R et al: MiR-122 promotes renal cancer cell proliferation by targeting Sprouty2. Tumor Biol 2017, 39(2): 1010428317691184.
- Jingushi K, Kashiwagi Y, Ueda Y, Kitae K, Hase H, Nakata W, Fujita K, Uemura M, Nonomura N, Tsujikawa K et al: High miR-122 expression promotes malignant phenotypes in ccRCC by targeting occludin. Int J Oncol 2017, 51(1): 289-297.
- Ji H, Tian D, Zhang B, Zhang Y, Yan D, Wu S: Overexpression of miR155 in clearcell renal cell carcinoma and its oncogenic effect through targeting FOXO3a. Exp Ther Med 2017, 13(5): 2286-2292.
- Hu G, Lai P, Liu M, Xu L, Guo Z, Liu H, Li W, Wang G, Yao X, Zheng J et al: miR-203a regulates proliferation, migration, and apoptosis by targeting glycogen synthase kinase-3β in human renal cell carcinoma. Tumor Biol 2014, 35(11): 11443-11453.
- He H, Dai J, Zhuo R, Zhao J, Wang H, Sun F, Zhu Y, Xu D: Study on the mechanism behind lncRNA MEG3 affecting clear cell renal cell carcinoma by regulating miR‐7/RASL11B signaling. J Cell Physiol 2018, 233(12): 9503-9515.
- Fan Y, Ma X, Li H, Gao Y, Huang Q, Zhang Y, Bao X, Du Q, Luo G, Liu K et al: miR‐122 promotes metastasis of clear‐cell renal cell carcinoma by downregulating Dicer. Int J Cancer 2018, 142(3): 547-560.
- Gong X, Zhao H, Saar M, Peehl DM, Brooks JD: miR-22 regulates invasion, gene expression and predicts overall survival in patients with clear cell renal cell carcinoma. Kidney Cancer 2019, 3(2): 119-132.
- Lv X, Shen J, Guo Z, Kong L, Zhou G, Ning H: Aberrant expression of miR-592 is associated with prognosis and progression of renal cell carcinoma. OncoTargets Ther 2019, 12: 11231-11239.
- Chi XG, Meng XX, Ding DL, Xuan XH, Chen YZ, Cai Q, Wang A: HMGA1-mediated miR-671-5p targets APC to promote metastasis of clear cell renal cell carcinoma through Wnt signaling. Neoplasma 2020, 67(1): 46-53.
- Wang J, Wang C, Li Q, Guo C, Sun W, Zhao D, Jiang S, Hao L, Tian Y, Liu S et al: miR-429-CRKL axis regulates clear cell renal cell carcinoma malignant progression through SOS1/MEK/ERK/MMP2/MMP9 pathway. Biomed Pharmacother 2020, 127: 110215.
- Li J, Jiang D, Zhang Q, Peng S, Liao G, Yang X, Tang J, Xiong H, Pang J: MiR-301a promotes cell proliferation by repressing PTEN in renal cell carcinoma. Cancer Manag Res 2020, 12: 4309-4320.
Annals of urologic oncology
p-ISSN: 2617-7765, e-ISSN: 2617-7773
 Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript