Review Article | Open Access
Advancing Genomics in Urologic Tumors: Navigating Precision Therapeutic Pathways
Fawad Inayat1, Imad Tariq2, Nabiha Bashir3, Fawad Ullah4, Hadiqa Aimen5
1Department of Biochemistry, Institute of Chemical and Life Sciences, Abdul Wali Khan University, Mardan, Pakistan.
2Department of Health and Biological Sciences, Abasyn University Peshawar, Pakistan.
3Khwaja Fareed University of Engineering & Information Technology (KFUEIT), Pakistan.
4Michigan Technological University, USA.
5COMSATS University Islamabad, Pakistan.
Correspondence: Fawad Inayat (Department of Biochemistry, Institute of Chemical and Life Sciences, Abdul Wali Khan University, Mardan, Pakistan; Email: fawad.313b@gmail.com).
Annals of Urologic Oncology 2024, 7(1): 33-42. https://doi.org/10.32948/auo.2024.05.18
Received: 02 May 2024 | Accepted: 18 May 2024 | Published online: 20 May 2024
Key words urothelial carcinoma, urological tumors, single nucleotide polymorphism (SNP) sites, transitional cell carcinoma and next-generation sequencing (NGS)
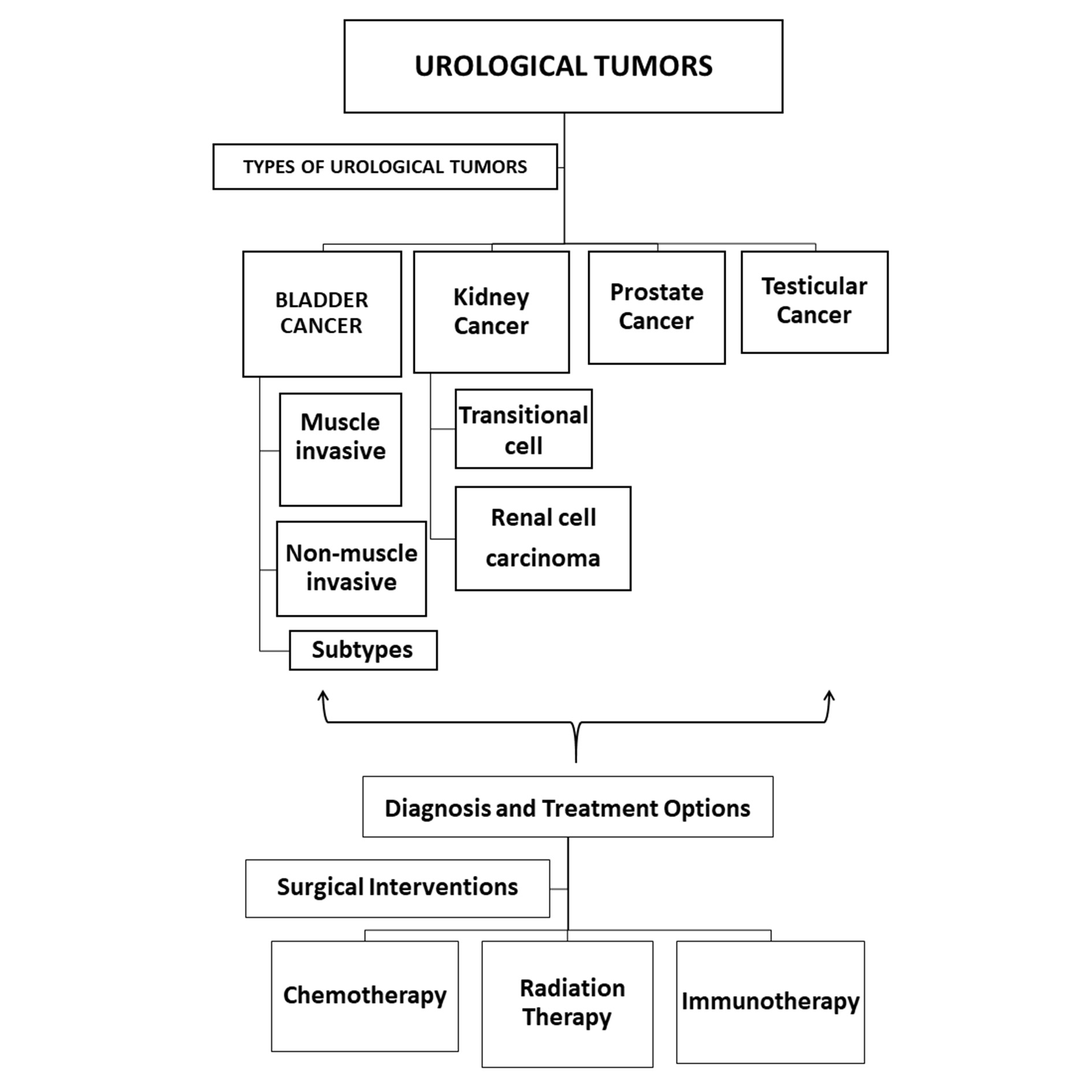 Figure 1. Illustration depicting various urological tumor types, diagnostic procedures, and treatment modalities, including surgery, chemotherapy, immunotherapy, and radiation therapy.
Figure 1. Illustration depicting various urological tumor types, diagnostic procedures, and treatment modalities, including surgery, chemotherapy, immunotherapy, and radiation therapy.
Recent advancements in genomics have enabled researchers to delve deep into the molecular mechanisms underlying urological cancers. Studies have revealed the complex interaction of genetic and cellular alterations that trigger the onset and advancement of urothelial, prostate, kidney, and testicular cancers. Understanding these underlying molecular mechanisms is crucial for developing targeted therapies that can effectively combat these diverse malignancies [5].
Deciphering bladder cancer molecular subtypes
In the realm of bladder cancer, researchers have made significant strides in deciphering the molecular subtypes that underlie its heterogeneous nature. Substantial efforts have been directed towards identifying distinct molecular subgroups within bladder cancer, each characterized by unique genetic alterations and molecular signatures. This molecular subtyping holds great promise for stratifying patients based on their specific tumor biology, ultimately aiding in the development of personalized treatment approaches [6].
Development of UBC diagnostic tool
The development of a reliable and accurate diagnostic tool for urothelial bladder cancer has been a focal point of research endeavors. Efforts have been geared towards harnessing genomic and molecular profiling to create a robust diagnostic tool that can effectively detect and classify UBC. Such a tool holds immense potential in facilitating early detection, guiding treatment decisions, and monitoring disease progression in patients with UBC [7].
Advancements in UBC diagnostics using methylation markers
Methylation markers have emerged as a promising avenue for advancing diagnostics in urothelial bladder cancer. These epigenetic modifications exhibit distinct patterns in UBC, serving as valuable biomarkers for early detection and prognostication. Integrating methylation markers into diagnostic protocols has the potential to enhance the precision and accuracy of UBC diagnosis, thus improving patient outcomes [8].
Epidemiology and risk factors in urological cancers
In addition to unraveling the molecular intricacies of urological cancers, epidemiological studies have shed light on the various risk factors associated with these malignancies. Environmental exposures, genetic predispositions, and lifestyle factors have been implicated in the etiology of bladder, prostate, kidney, and testicular cancers. Understanding the epidemiology and risk factors is integral in devising comprehensive prevention and early detection strategies to mitigate the burden of urological cancers on a global scale [9].
This review is connecting key aspects of urological cancer research, as illustrated in the Figure 2 intricate relationship between molecular mechanisms, bladder cancer subtyping, diagnostic tool innovation, and methylation marker advancements. It serves as a comprehensive visual guide, highlighting the multifaceted approach to understanding and managing urological tumors and their types.
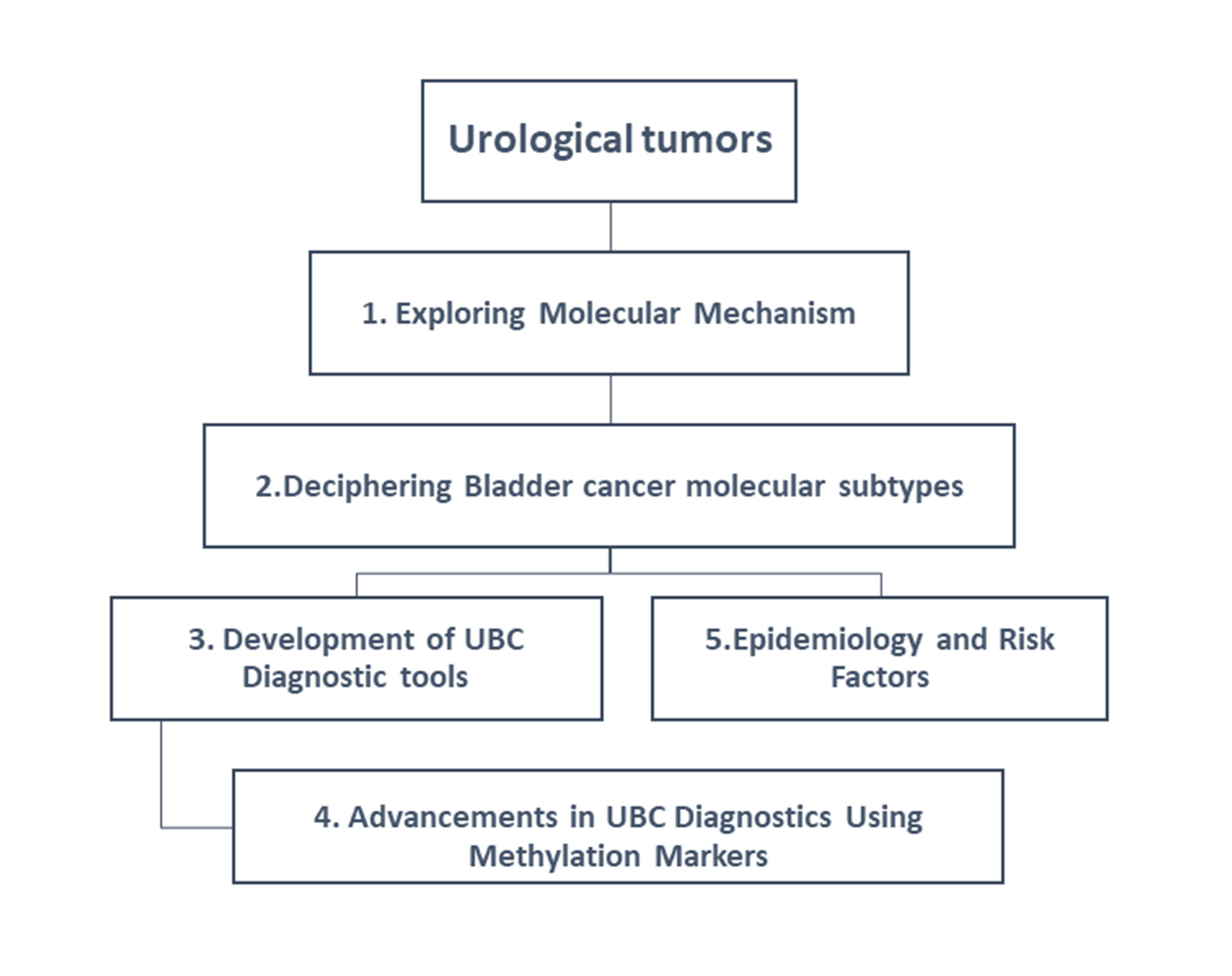 Figure 2. Mapping the interconnections between urological cancer research components and tumor types.
Figure 2. Mapping the interconnections between urological cancer research components and tumor types.
The importance of genomic profiling in urological tumors
Urological cancers, encompassing malignancies of the bladder, prostate, kidney, and testes, represent a significant global health challenge with increasing incidence rates and associated morbidity and mortality. Among these, transitional cell cancer stands out as the dominant tissue-based type of bladder cancer, presenting a complex molecular landscape that drives cancer progression. This delves into recent genomic research that unravels the intricate DNA-related and epigenomic changes underpinning the advancement of urological malignancies, shedding light on the diverse genetic landscape and molecular subtypes of bladder cancer. The study emphasizes the pivotal role of epigenomic control proteins and their counterparts in cancer advancement, highlighting how aberrant activity can lead to gene expression dysregulation and promote tumorigenesis. Furthermore, it details the unique genetic alterations observed in urothelial cancers, differentiating between papillary and non-papillary subtypes and their implications for prognosis and treatment strategies. By understanding the genetic and molecular characteristics of these urological malignancies, clinicians and researchers can tailor precision therapeutic pathways to the individual needs of patients. This individualized method shows significant potential in enhancing treatment results and overall well-being of patients [3, 12]. Furthermore, the integration of genomic and molecular profiling into diagnostic protocols can facilitate early detection, guide treatment decisions, and monitor disease progression, thus contributing to more effective management of urological cancers.
Advancing our understanding of urologic tumors by delving into their genetic and molecular characteristics is paramount in the pursuit of tailored precision therapies. The diversity and clinical complexity that these tumors present necessitate a deep exploration of their genomic makeup. Genomic profiling holds the key to unlocking personalized treatment pathways, ultimately catering to the unique needs of each patient. By unraveling the intricate genetic and molecular landscape of urologic tumors, we can navigate towards enhanced therapeutic strategies that offer improved outcomes and quality of life for patients [13].
Bladder cancer, particularly urothelial carcinoma, stands out as a predominant and clinically complex subtype, contributing substantially to the global burden of cancer-related morbidity and mortality. It is essential to continue exploring precision therapeutic pathways to address the challenges posed by urologic cancers and advance genomics in the understanding and treatment of these tumors. This progress can pave way for more precise and competent treatments tailored to the specific genomic characteristics of urologic tumors, ultimately improving patient outcomes and quality of life [14]. Notably urothelial carcinoma ranks as the fourth most frequently diagnosed cancer in men and the eighth in women [15].
The study emphasizes the pivotal role of epigenomic controlled proteins and their counterparts in cancer advancement, highlighting how aberrant activity can lead to gene expression dysregulation and promote tumorigenesis. Furthermore, it details the unique genetic alterations observed in urothelial cancers, differentiating between papillary tumors and invasive carcinomas. Mutations in genes like FGFR3, HRAS, and PIK3CA characterize papillary tumors, while the loss of chromosome 9 targeting CDKN2A and TSC1 is prominent in invasive carcinomas. This distinction underscores the importance of understanding the cellular heterogeneity of urological tumor to tailor management strategies effectively. In addition to genetic alterations, the study explores the significance of transcription factors such as E2F1 and E2F3 in driving cell proliferation and replication in invasive bladder cancers. It emphasizes the critical role of the RB1 protein in restraining these factors, and its dysregulation in bladder cancer leads to uncontrolled cell growth and tumorigenesis. Understanding the interplay between epigenetic regulatory proteins, genetic alterations, and transcription factors is crucial in comprehensively addressing the molecular intricacies of bladder cancer [16]. By delving into these molecular mechanisms, researchers and clinicians can develop targeted therapies that specifically modulate the dysregulated pathways, ultimately improving treatment efficacy and patient outcomes. The integration of these findings into diagnostic and therapeutic paradigms holds immense promise in advancing personalized medicine for bladder cancer. Tailoring treatment approaches based on the molecular characteristics of individual tumors can significantly enhance the precision and effectiveness of interventions, ultimately improving the prognosis and quality of life for patients [17].
Understanding the heterogeneity of bladder cancer
Transitional cell carcinoma of the bladder, a varied disease with diverse clinical presentations and outcomes, poses a significant challenge in oncology. The study titled "Genomic Subtyping of Bladder Cancer and its Clinical Implications" delves into the cellular intricacies of urothelial cancer, emphasizing the importance of genomic and transcriptomic profiling in understanding disease heterogeneity and guiding personalized treatment strategies [18]. Spatial tumor heterogeneity further complicates the landscape of bladder cancer, highlighting the necessity for comprehensive molecular characterization to unravel the disease's complex genetic architecture. The study provides a detailed exploration of the molecular subtypes across early stage bladder cancer and advanced bladder cancer. The study identified distinct molecular variations within NMIBC, associating specific characteristics like luminal papillary, genomically unstable, and basal-like features with different subtypes.In addition to unraveling the molecular intricacies, it is crucial to understand the implications of these findings on the medical therapy of urothelial tumor. The identified molecular subtypes have the capacity to transform treatment strategies by facilitating tailored and individualized approaches that consider the specific genetic characteristics of each subtype [19]. Moving forward, the integration of molecular subtyping and genomic profiling into diagnostic and treatment protocols can significantly impact the administration of urothelial bladder tumor, leading to more precise and effective personalized therapies for patients. This comprehensive knowledge of the cellular heterogeneity of urothelial cancer paves the way for improved patient outcomes and a deeper exploration of precision medicine in the field of oncology [20]. Delving into the molecular heterogeneity of bladder cancer, this study presents a meticulous analysis of its various subtypes and their clinical implications. Utilizing advanced genomic and transcriptomic methodologies, the study delineates the multifaceted genetic landscape of bladder cancer. By identifying distinct molecular subtypes, the study provides critical insights that could revolutionize treatment approaches, paving the way for personalized therapeutic interventions tailored to individual patients' genetic profiles.Understanding the molecular subtypes of bladder cancer is crucial for refining treatment strategies and improving patient outcomes. With the intricate genetic landscape of bladder cancer unveiled, targeted and effective treatments can be developed to address the specific characteristics of each subtype. This approach holds the promise of enhancing therapeutic outcomes and elevating well-being of individuals of those impacted. As research continues to uncover the complexities of bladder cancer at the molecular level, the potential for tailored precision therapies becomes increasingly promising. The identification of molecular subtypes opens new avenues for personalized treatment pathways, offering renewed hope for individuals battling this clinically complex malignancy [21]. Bladder cancer manifests across a broad clinical spectrum, encompassing early-stage urothelial bladder cancer and advanced urothelial bladder cancer, each with distinct biological behaviors, treatment paradigms, and prognostic implications. While NMIBC often recurs but rarely progresses to invasive disease, MIBC, characterized by infiltration beyond the muscularis propria, carries a heightened risk of metastasis and mortality, necessitating aggressive therapeutic interventions.Understanding these distinctions is crucial in developing precision therapeutic pathways for bladder cancer. By leveraging genomics, researchers and clinicians can identify specific genetic and molecular characteristics that differentiate NMIBC and MIBC, leading to the development of targeted therapies that address the unique biological behaviors of each subtype.Advancing genomics in the understanding and treatment of urothelial tumor holds the capacity to change patient care by enabling the delivery of personalized, precision medicine. This approach can significantly improve the effectiveness of treatments while minimizing adverse effects, ultimately leading to better outcomes and quality of life for individuals affected by bladder cancer [22].
Bladder cancer manifests through two distinct pathways: non-muscle-invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer. In NMIBC, common genetic alterations include the deletion of chromosome 9 and mutations in fibroblast growth factor receptor (FGFR) genes, notably FGFR3. These alterations are often observed in hyperplastic precursor lesions, indicating their role in early bladder cancer development. Conversely, muscle-invasive bladder cancer presents a different molecular profile characterized by more aggressive genetic changes. While chromosome 9 deletion and FGFR mutations may still occur, additional alterations in genes such as TP53, RB1, and PIK3CA are commonly detected. These genetic changes propel tumor progression, leading to invasion into the muscle layer of the bladder wall and potential metastasis. Understanding the distinct genetic and molecular alterations associated with each pathway is crucial for accurate diagnosis, prognosis, and treatment selection in bladder cancer patients. Detecting these alterations in precursor lesions holds promise for early intervention and preventive measures to mitigate disease progression. Overall, unraveling the complexities of bladder cancer pathways, including the genetic and molecular changes involved, is imperative for advancing our understanding of the disease and developing targeted therapeutic strategies to enhance patient outcomes are given ( in Figure 3).
Development of UBC diagnostic tool
The exploration of bladder cancer's molecular subtypes and understanding their experimental significance is important for tailoring treatment strategies and optimizing patient outcomes [23]. A retrospective analysis revealed that tumors with heightened immune activation correlated with superior 5-year disease-specific survival rates, suggesting a potential avenue for further research.The study dives into the molecular intricacies of early-stage urothelial bladder cancer and advanced urothelial bladder cancer, identifying distinct molecular subtypes with specific genetic characteristics [24]. This highlights the necessity for detailed molecular profiling to decipher the disease's complex genetic landscape and underscores the potential for personalized therapeutic interventions tailored to individual patients' genetic profiles [25].
The integration of molecular subtyping and genomic profiling into diagnostic and treatment protocols holds significant promise for enhancing the management of bladder cancer.This groundbreaking study aims to revolutionize bladder cancer diagnostics by harnessing the potential of methylation markers and single nucleotide polymorphism sites [26]. Leveraging cutting-edge next-generation sequencing technologies, sophisticated bioinformatics algorithms, and rigorous validation protocols, the study meticulously identifies potential diagnostic biomarkers associated with urothelial bladder carcinoma. The development of this precise diagnostic tool holds immense promise in enhancing early detection rates, facilitating timely interventions, and ultimately improving patient outcomes [27] .
By delving into the cellular landscape of urothelial cancer and harnessing the power of genomic profiling, this study seeks to pave the way for the development of a highly accurate and targeted diagnostic tool. The utilization of methylation markers and SNP sites, in combination with advanced NGS technologies and bioinformatics algorithms, aims to identify specific biomarkers that can aid in the early and precise detection of UBC [28]. The successful development and implementation of this diagnostic tool have the potential to transform the clinical management of UBC, offering healthcare professionals a sophisticated and reliable method for detecting and diagnosing this complex malignancy [29]. Ultimately, this innovation has the capacity to significantly improve patient outcomes by enabling timely interventions and tailored treatment strategies.The etiological landscape of bladder cancer is multifaceted, shaped by a complex interplay of genetic predispositions, environmental exposures, and lifestyle factors. Risk variables like tobacco use and exposure to workplace carcinogens, chronic bladder inflammation, along with genetic mutations have been identified as significant contributors to bladder cancer initiation and progression [30]. Understanding the intricate relationship between these risk factors and the genetic and molecular characteristics of bladder cancer is pivotal in developing precision therapeutic pathways tailored to the individual needs of patients. By delving into the genetic underpinnings of bladder cancer and considering the influence of environmental and lifestyle factors, researchers and clinicians can enhance the development of targeted therapies that address the complexities of this disease. This comprehensive approach holds the promise of optimizing treatment outcomes.
Advancements in UBC diagnostics using methylation markers
With a focus on redefining diagnostic paradigms for urothelial bladder carcinoma, this study explores the transformative potential of methylation markers and SNP sites in enhancing diagnostic accuracy. Through a comprehensive approach that integrates NGS, bioinformatics analysis, and meticulous validation studies, the study identifies and authenticates novel diagnostic biomarkers [31]. These findings not only provide unprecedented insights into the pathophysiology of UBC but also herald a new era of targeted diagnostic strategies, offering renewed hope for early detection and effective management of this aggressive malignancy.The utilization of methylation markers and SNP sites, in combination with advanced NGS technologies and bioinformatics algorithms, aims to identify specific biomarkers that can aid in the early and precise detection of UBC [32]. This symbolizes a significant leap onward in the development of diagnostic tools for UBC, with the potential to revolutionize clinical practices and significantly improve patient outcomes. By incorporating these advancements into the diagnostic process, healthcare professionals can offer more targeted and timely interventions, ultimately leading to improved prognosis and management of UBC [33]. The successful implementation of these advanced diagnostic approaches has the potential to transform the landscape of clinical care for individuals with UBC, providing a much-needed breakthrough in early detection and personalized treatment strategies. As research in this area continues to progress, the integration of methylation markers and SNP sites into diagnostic protocols is poised to become a cornerstone of UBC management, offering renewed hope and improved care for patients affected by this challenging malignancy [34]. As the global population ages and environmental exposures continue to evolve, understanding the epidemiology, risk factors, and early detection strategies for bladder cancer becomes increasingly imperative [35].
Epidemiology and risk factors in urological cancers
Bladder cancer, renal cell carcinoma, and testicular cancer are significant urological malignancies with distinct epidemiological profiles and risk factors [36]. The profound impact of environmental exposures, gender disparities, and lifestyle factors on the incidence and outcomes of these cancers cannot be overstated.Bladder cancer stands as a significant global health challenge, with its incidence closely tied to modifiable and non-modifiable risk factors [37]. Smoking remains a predominant risk factor, with the consumption of aristolochic acid-containing Chinese herbal medicines also contributing to the increased prevalence of upper tract urothelial carcinoma [38].
Bladder cancer is known for its multifaceted etiological landscape, with risk variables like tobacco use and exposure to workplace carcinogens, chronic bladder inflammation, and genetic mutations playing pivotal roles in its initiation and progression. Understanding the intricate relationship between these risk factors and the genetic and molecular characteristics of bladder cancer is critical in developing precision therapeutic pathways tailored to individual patient needs [39].
Prostate cancer has an intricate genetic profile that is essential in the development and advancement of the illness. Identifying and describing changes in prostate cancer genes may provide a critical understanding of the underlying molecular processes that cause the disease. The TMPRSS2-ERG gene fusion is a prominent genetic mutation seen in prostate cancer, present in about 50% of cases. The fusion occurs due to the translocation between the TMPRSS2 gene, controlled by androgens, and the ERG gene, which belongs to the ETS transcription factor family. The excessive production of ERG, controlled by androgen signalling, leads to the development of cancer by disrupting the regulation of cell division, facilitating the spread of cancer cells, and affecting the surrounding environment of the tumour. Furthermore, a proportion of prostate tumours, those categorized as metastatic castration-resistant prostate cancers (mCRPC), have mutations in DNA repair genes such as BRCA1, BRCA2, and ATM. These abnormalities hinder the homologous recombination repair (HRR) mechanism, crucial for fixing double-strand DNA breaks. As a result, these mutations make individuals more vulnerable to some therapeutic drugs. Identifying HRR pathway defects has accelerated the development of tailored treatment approaches, particularly poly (ADP-ribose) polymerase (PARP) inhibitors. PARP inhibitors, such as olaparib and rucaparib, use synthetic lethality by further impairing the ability of cancer cells to repair DNA, which is already compromised owing to BRCA1/2 mutations. PARP inhibitors are very effective in clinical studies, increasing the amount of time that mCRPC patients with these mutations may live without their disease progressing. This highlights the need to use genomic profiling to help decide treatment options. Furthermore, identifying TMPRSS2-ERG fusions has stimulated research on explicitly targeting the ERG protein and studying the consequences of its excessive expression. While there are currently no therapeutically accessible drugs that directly suppress ERG, current research focuses on disrupting the signalling pathways regulated by ERG, such as those involving PI3K/Akt and MAPK. This approach may provide new opportunities for intervention. Advanced diagnostic tools, such as next-generation sequencing and liquid biopsies, make incorporating genetic changes into clinical treatment easier. These approaches provide extensive tumour information and may identify minimal residual illness or growing resistance. This feature allows medical professionals to customize therapies more accurately and modify therapeutic approaches based on the tumour's genetic profile as it evolves.
Renal cancer, also known as renal cell carcinoma (RCC), is characterized by a wide range of genetic changes that have a substantial impact on its development, clinical course, and response to treatment. Renal cancer exhibits genetic diversity, with clear cell RCC (ccRCC) being the most prevalent subtype. This subtype is distinguished by distinct genetic changes that contribute to its pathogenesis. The presence of a mutation in the von Hippel-Lindau (VHL) tumour suppressor gene is a defining characteristic of the ccRCC cancer subtype, occurring in around 70% of cases. The loss of VHL function results in the buildup of hypoxia-inducible factors (HIFs), which then triggers the activation of angiogenic, proliferative, and metastatic pathways via the mediation of vascular endothelial growth factor (VEGF) and other growth factors. Furthermore, modifications in several additional genes are crucial in developing RCC. Approximately 40% of clear renal cell carcinoma (ccRCC) patients have polybromo-1 (PBRM1) gene mutations. These mutations lead to changes in chromatin remodelling and the control of gene expression. SETD2, BAP1, and KDM5C have often altered genes that contribute to cancer's genomic instability and aggressive behaviour. They do so via different methods that include modifying histones and repairing DNA. The comprehension of these genetic changes has significantly influenced the treatment options available for kidney cancer. Therapies targeting the VEGF pathway, including sunitinib and pazopanib, have been created by understanding the processes caused by VHL mutation and HIF buildup. These medications have shown substantial effectiveness in enhancing the survival rates of patients with RCC, especially in the later stages of the illness. Moreover, identifying mutations in genes involved in chromatin remodeling, such as PBRM1 and BAP1, has provided new opportunities for developing therapeutic approaches that specifically target epigenetic regulators. Current research assesses the effectiveness of inhibitors that target these pathways, possibly providing novel therapeutic choices for individuals with specific genetic profiles. Integrating extensive genetic profiling into the clinical management of kidney cancer enables a more sophisticated therapy strategy. Advanced genomic testing, such as whole-exome and targeted gene sequencing, provides essential information on the precise genetic changes in a tumour. This information helps choose appropriate targeted treatments and supports a precision medicine strategy. Furthermore, the growing use of liquid biopsies in identifying circulating tumour DNA presents a hopeful means of continuously monitoring the progression of tumours and their resistance to treatment. This enables prompt modifications in treatment approaches to effectively meet the ever-changing characteristics of cancer genetics.
Testicular cancer, although less common, presents its own set of risk factors, including cryptorchidism, family history, and genetic abnormalities. Understanding these factors is crucial in devising effective screening programs and preventive initiatives for this particular urological cancer [40]. The comprehensive understanding of epidemiological data and risk factors for these urological cancers underscores the imperative for targeted prevention strategies, robust screening programs, and public health initiatives aimed at mitigating the burden of these malignancies and improving population-wide health outcomes [41]. These efforts are essential in addressing the increasing prevalence of urological cancers in our aging population and in adapting to evolving environmental exposures [42]. Recent advancements in genomic and molecular research have revolutionized our understanding of bladder cancer biology, unveiling intricate molecular pathways and genetic and epigenetic alterations driving tumorigenesis, progression, and therapeutic resistance [43]. Comprehensive genomic and transcriptomic analyses have identified distinct molecular subtypes, genetic mutations, copy number alterations, and epigenomic changes associated with urothelial cancer, elucidating the diversity and complexity of the disease [44]. Incorporating this knowledge into precision therapeutic pathways holds significant promise for improving patient outcomes and quality of life.
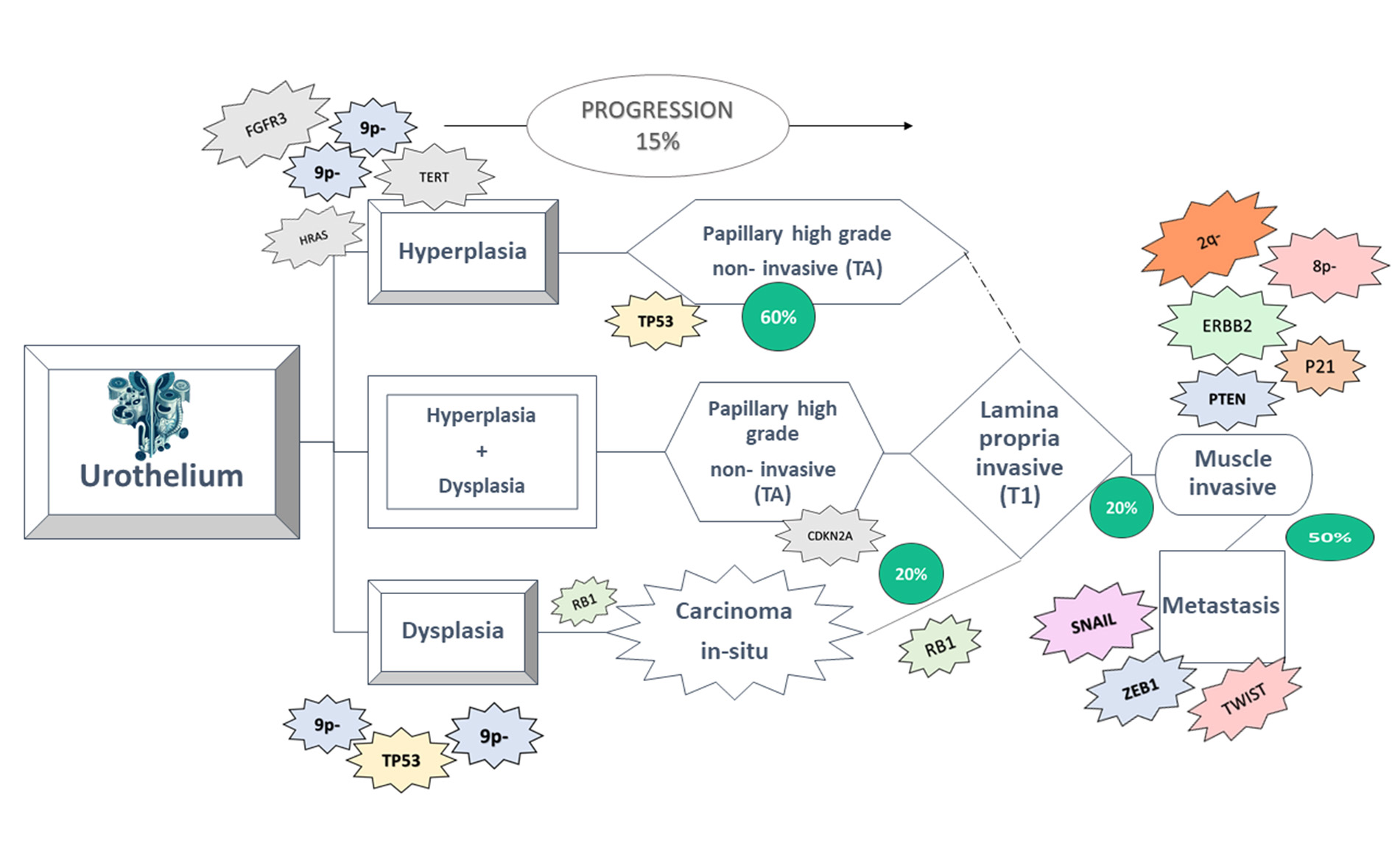 Figure 3. Bladder cancer progresses through two separate routes, resulting in the development of non-muscle-invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer. Changes often found in NMIBC include chromosome 9 deletion and FGFR point mutation, which are also observed in hyperplastic precursor cells [22].
Figure 3. Bladder cancer progresses through two separate routes, resulting in the development of non-muscle-invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer. Changes often found in NMIBC include chromosome 9 deletion and FGFR point mutation, which are also observed in hyperplastic precursor cells [22].
The rise of next-generation sequencing has revolutionized DNA methylation profiling, enabling researchers to explore epigenetic modifications with incredible precision. These advanced techniques provide in-depth analysis of DNA methylation patterns throughout the genome, enhancing our understanding of their role in gene regulation, development, and disease. One of the powerful techniques used for DNA methylation profiling is whole-genome bisulfite sequencing, which provides single-nucleotide resolution of DNA methylation across the entire genome. By treating DNA with bisulfite, which converts unmethylated cytosines to uracil, WGBS allows for the identification of methylated cytosines at a genome-wide scale. Additionally, reduced representation bisulfite sequencing is another method that focuses on sequencing a subset of genomic regions enriched for CpG sites, providing a cost-effective approach for DNA methylation profiling. These techniques have significantly contributed to our understanding of the complex regulatory mechanisms mediated by DNA methylation. Moreover, recent advancements in NGS have led to the development of targeted bisulfite sequencing, which allows for the selective analysis of specific genomic regions of interest, offering a more efficient and economical alternative to whole-genome bisulfite sequencing. This targeted approach is particularly advantageous for studying DNA methylation in specific gene promoters or regulatory elements. Another emerging technique, known as single-cell bisulfite sequencing, has enabled the assessment of DNA methylation patterns at the single-cell level, providing insights into the heterogeneity of methylation profiles within a population of cells.Advancements in next-generation sequencing (NGS) have revolutionized DNA methylation profiling, equipping researchers with sophisticated tools to investigate epigenetic modifications with unparalleled precision (given in Figure 4). These techniques facilitate comprehensive analysis of DNA methylation patterns across the genome, deepening our understanding of their roles in gene regulation, development, and disease [51].
1. BS-Seq (Bisulfite Sequencing):Utilizes bisulfite treatment to convert unmethylated cytosines to uracils, allowing for high-resolution mapping of DNA methylation patterns.
2. MeDIP-Seq (Methylated DNA Immunoprecipitation Sequencing):Relies on specific antibodies to enrich methylated regions for sequencing, offering cost-effective profiling of DNA methylation patterns with relatively lower resolution.
3. RRBS-Seq (Reduced Representation Bisulfite Sequencing):Combines bisulfite treatment with restriction enzyme digestion to focus on CpG-rich regions, providing efficient coverage of regulatory elements at reduced sequencing costs.
4. WGBS (Whole Genome Bisulfite Sequencing): Analyzes DNA methylation across the entire genome without prior digestion, yielding comprehensive methylation profiles albeit requiring deeper sequencing coverage.
5. MethylCap-Seq (Methylation Capture Sequencing): Captures methylated DNA fragments using methyl-CpG binding proteins, enhancing sensitivity and specificity of methylation detection.
6. MBD-Seq (Methyl-CpG Binding Domain Sequencing): Selectively enriches methylated DNA with methyl-CpG binding proteins, striking a balance between specificity and genome coverage.
7. oxBS.Seq (Oxidative Bisulfite Sequencing): Distinguishes between 5mC and 5hmC by combining bisulfite treatment with oxidation, providing insights into both DNA methylation and hydroxymethylation patterns.
8. TAB-Seq (TET-associated Bisulfite Sequencing):Profiles DNA methylation and hydroxymethylation simultaneously by oxidizing 5mC to 5hmC with TET proteins, followed by bisulfite treatment and sequencing.
9. BSAS (Bisulfite Amplicon Sequencing): Enables quantitative analysis of DNA methylation at specific loci through bisulfite treatment and targeted PCR amplification, useful for targeted methylation studies and validating genome-wide findings [52].
Advances in next-generation sequencing (NGS) have significantly transformed bladder cancer research, especially in understanding its epigenetic nuances. Methods like BS-Seq, MeDIP-Seq, and WGBS allow for detailed examination of DNA methylation patterns, revealing possible biomarkers and treatment targets. This thorough analysis unveils abnormal methylation occurrences critical for bladder cancer progression, providing insights into its underlying molecular mechanisms. Through the characterization of these epigenetic alterations, NGS techniques offer potential for tailored diagnostic approaches and personalized therapies, heralding a new era in managing bladder cancer.
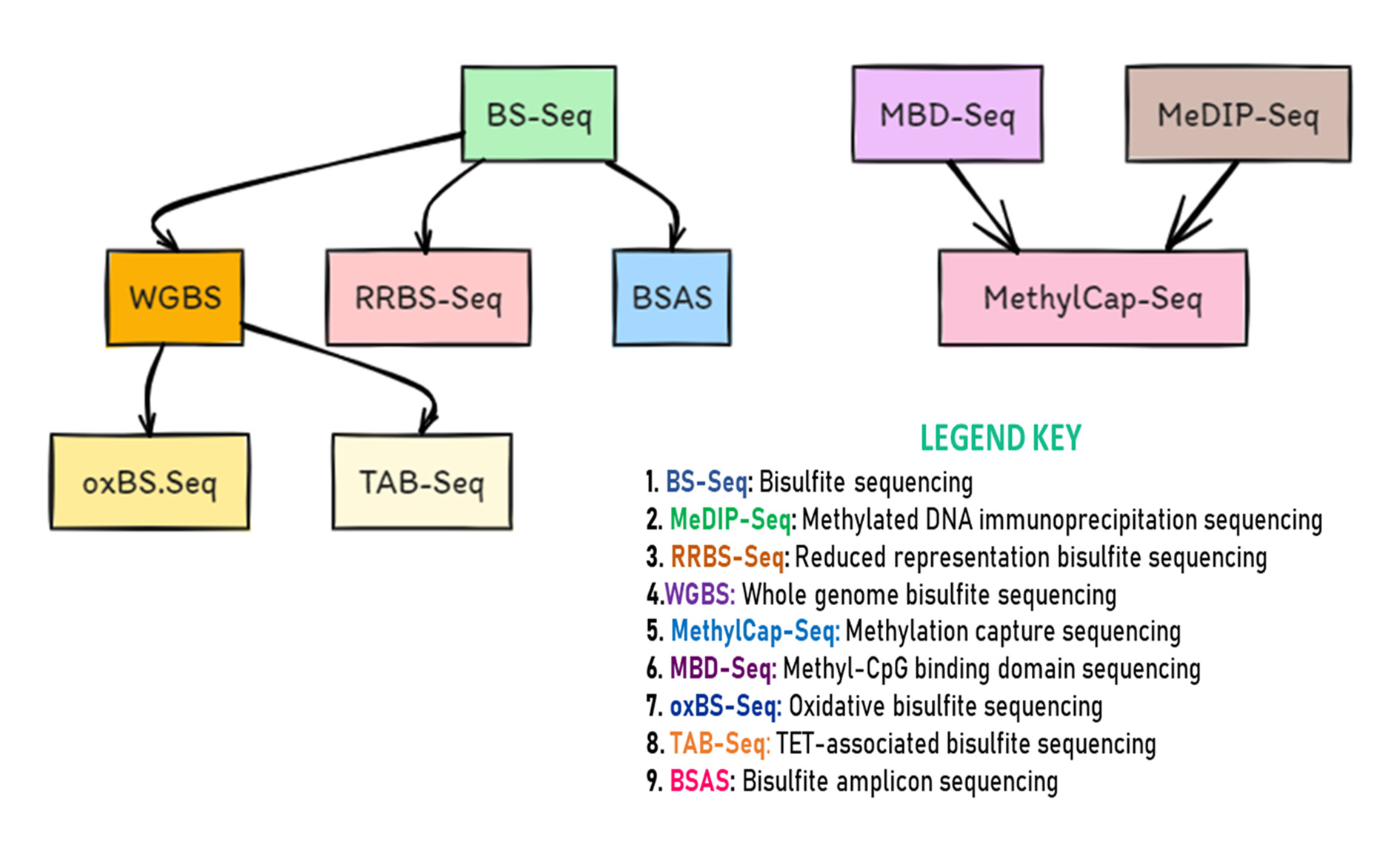 Figure 4. Next-Generation Sequencing Methods for DNA Methylation Analysis.
Figure 4. Next-Generation Sequencing Methods for DNA Methylation Analysis.
None.
Ethical policy
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study. Approval from institutional ethical committee was taken.
Availability of data and materials
All data generated or analysed during this study are included in this publication.
Author contributions
FI, IT: Conception, design of study, literature search and review, manuscript writting; NB, FU: figure production; HA: Supervision and approval for the final version of the manuscript.
Competing interests
The authors have no competing interest.
Funding
None.
- Zaghloul MS: Bladder cancer and schistosomiasis. J Egypt Natl Canc Inst 2012, 24(4): 151-159.
- Mazzaschi G, Giudice GC, Corianò M, Campobasso D, Perrone F, Maffezzoli M, Testi I, Isella L, Maestroni U, Buti S: Upper Tract Urinary Carcinoma: A Unique Immuno-Molecular Entity and a Clinical Challenge in the Current Therapeutic Scenario. Technol Cancer Res Treat 2023, 22: 15330338231159753.
- McConkey DJ: Molecular Biology of Bladder Cancer: Potential Implications for Therapy. Hematol Oncol Clin North Am 2021, 35(3): 457-468.
- Droller MJJTJoU: Urological Oncology: Bladder, Penis and Urethra Cancer, and Basic Principles of Oncology. J Urol 2003, 170(5): 2116-2125.
- Tran L, Xiao JF, Agarwal N, Duex JE, Theodorescu D: Advances in bladder cancer biology and therapy. Nat Rev Cancer 2021, 21(2): 104-121.
- Li HT, Duymich CE, Weisenberger DJ, Liang G: Genetic and Epigenetic Alterations in Bladder Cancer. Int Neurourol J 2016, 20(Suppl 2): S84-94.
- Wang Y, Chen L, Ju L, Qian K, Liu X, Wang X, Xiao Y: Novel Biomarkers Associated With Progression and Prognosis of Bladder Cancer Identified by Co-expression Analysis. Front Oncol 2019, 9: 1030.
- Zhou Q, Chen X, Yao K, Zhang Y, He H, Huang H, Chen H, Peng S, Huang M, Cheng L et al: TSPAN18 facilitates bone metastasis of prostate cancer by protecting STIM1 from TRIM32-mediated ubiquitination. J Exp Clin Cancer Res 2023, 42(1): 195.
- Saginala K, Barsouk A, Aluru JS, Rawla P, Padala SA, Barsouk A: Epidemiology of Bladder Cancer. Med Sci (Basel) 2020, 8(1): 15.
- Choi W, Ochoa A, McConkey DJ, Aine M, Höglund M, Kim WY, Real FX, Kiltie AE, Milsom I, Dyrskjøt L et al: Genetic Alterations in the Molecular Subtypes of Bladder Cancer: Illustration in the Cancer Genome Atlas Dataset. Eur Urol 2017, 72(3): 354-365.
- Dy GW, Gore JL, Forouzanfar MH, Naghavi M, Fitzmaurice C: Global Burden of Urologic Cancers, 1990-2013. Eur Urol 2017, 71(3): 437-446.
- McConkey DJ, Singla N, Pierorazio P, Lombardo K, Matoso A, Hoffman-Censits J: Molecular subtypes of upper tract urothelial cancer: Setting the stage for precision therapy. Cancer Cell 2021, 39(6): 745-747.
- Shen C, Fu C, Suo Y, Li K, Zhang Z, Yang S, Zhang Y, Lin Y, Li Z, Wu Z et al: Pan-cancer analyses of clinical prognosis, immune infiltration, and immunotherapy efficacy for TRPV family using multi-omics data. Heliyon 2023, 9(6): e16897.
- Schulz WA, Ribarska T: Insights into cancer mechanisms from genomic research on urological cancers. Genome Med 2011, 3(3): 20.
- Metts MC, Metts JC, Milito SJ, Thomas CR: Bladder cancer: a review of diagnosis and management. J Natl Med Assoc 2000, 92(6): 285-294.
- Kukkonen K, Taavitsainen S, Huhtala L, Uusi-Makela J, Granberg KJ, Nykter M, Urbanucci A: Chromatin and Epigenetic Dysregulation of Prostate Cancer Development, Progression, and Therapeutic Response. Cancers (Basel) 2021, 13(13): 3325.
- Chou R, Gore JL, Buckley D, Fu R, Gustafson K, Griffin JC, Grusing S, Selph S: Urinary Biomarkers for Diagnosis of Bladder Cancer: A Systematic Review and Meta-analysis. Ann Intern Med 2015, 163(12): 922-931.
- Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, Hinoue T, Laird PW, Hoadley KA, Akbani R et al: Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017, 171(3): 540-556.e525.
- Lerner SP, McConkey DJ, Hoadley KA, Chan KS, Kim WY, Radvanyi F, Höglund M, Real FX: Bladder Cancer Molecular Taxonomy: Summary from a Consensus Meeting. Bladder Cancer 2016, 2(1): 37-47.
- Linares-Espinós E, Sánchez-Salas R: Re: Robot-assisted Radical Cystectomy Versus Open Radical Cystectomy in Patients with Bladder Cancer (RAZOR): An Open-label, Randomised, Phase 3, Non-inferiority Trial. Eur Urol 2019, 75(1): 199.
- Guo CC, Shen SS, Ro JY: Pathogenesis and Diagnosis of Genitourinary Cancer. Cancers (Basel) 2021, 13(2): 347.
- Lopez-Beltran A, Cookson MS, Guercio BJ, Cheng L: Advances in diagnosis and treatment of bladder cancer. BMJ 2024, 384: e076743.
- Lindskrog SV, Prip F, Lamy P, Taber A, Groeneveld CS, Birkenkamp-Demtröder K, Jensen JB, Strandgaard T, Nordentoft I, Christensen E et al: An integrated multi-omics analysis identifies prognostic molecular subtypes of non-muscle-invasive bladder cancer. Nat Commun 2021, 12(1): 2301.
- Kamoun A, de Reyniès A, Allory Y, Sjödahl G, Robertson AG, Seiler R, Hoadley KA, Groeneveld CS, Al-Ahmadie H, Choi W et al: A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur Urol 2020, 77(4): 420-433.
- Asleh K, Negri GL, Spencer Miko SE, Colborne S, Hughes CS, Wang XQ, Gao D, Gilks CB, Chia SKL, Nielsen TO et al: Proteomic analysis of archival breast cancer clinical specimens identifies biological subtypes with distinct survival outcomes. Nat Commun 2022, 13(1): 896.
- Lasseigne BN, Burwell TC, Patil MA, Absher DM, Brooks JD, Myers RM: DNA methylation profiling reveals novel diagnostic biomarkers in renal cell carcinoma. BMC Med 2014, 12: 235.
- Bivalacqua TJ: Relationship Between the Number of Lymph Nodes Dissected and Prognosis in Patients With MIBC in the Era of Neoadjuvant Chemotherapy. Int J Urol, Epub ahead of print.
- Hong M, He G, Goh S, Low AWX, Tay KJ, Lim TKH, Yeong J, Khor LY, Lim TS: Biomarkers for Precision Urothelial Carcinoma Diagnosis: Current Approaches and the Application of Single-Cell Technologies. Cancers (Basel) 2021, 13(2): 260.
- Olkhov-Mitsel E, Savio AJ, Kron KJ, Pethe VV, Hermanns T, Fleshner NE, van Rhijn BW, van der Kwast TH, Zlotta AR, Bapat B: Epigenome-Wide DNA Methylation Profiling Identifies Differential Methylation Biomarkers in High-Grade Bladder Cancer. Transl Oncol 2017, 10(2): 168-177.
- Teoh JY: Epidemiology and screening for urologic cancers. World J Urol 2023, 41(4): 897-898.
- Peng D, Ge G, Xu Z, Ma Q, Shi Y, Zhou Y, Gong Y, Xiong G, Zhang C, He S et al: Diagnostic and prognostic biomarkers of common urological cancers based on aberrant DNA methylation. Epigenomics 2018, 10(9): 1189-1199.
- Köhler CU, Walter M, Lang K, Plöttner S, Roghmann F, Noldus J, Tannapfel A, Tam YC, Käfferlein HU, Brüning T: In-Vitro Identification and In-Vivo Confirmation of DNA Methylation Biomarkers for Urothelial Cancer. Biomedicines 2020, 8(8): 233.
- Shi ZD, Han XX, Song ZJ, Dong Y, Pang K, Wang XL, Liu XY, Lu H, Xu GZ, Hao L et al: Integrative multi-omics analysis depicts the methylome and hydroxymethylome in recurrent bladder cancers and identifies biomarkers for predicting PD-L1 expression. Biomark Res 2023, 11(1): 47.
- Yu Z, Lu C, Lai Y: A serum miRNAs signature for early diagnosis of bladder cancer. Ann Med 2023, 55(1): 736-745.
- Zhang J, Xu R, Lu Q, Xu Z, Liu J, Li P, Zhang Y, Zhou C, Luo L, Tang W et al: A Novel Methylation Marker NRN1 plus TERT and FGFR3 Mutation Using Urine Sediment Enables the Detection of Urothelial Bladder Carcinoma. Cancers (Basel) 2023, 15(3): 615.
- Jubber I, Ong S, Bukavina L, Black PC, Compérat E, Kamat AM, Kiemeney L, Lawrentschuk N, Lerner SP, Meeks JJ et al: Epidemiology of Bladder Cancer in 2023: A Systematic Review of Risk Factors. Eur Urol 2023, 84(2): 176-190.
- Cumberbatch MGK, Jubber I, Black PC, Esperto F, Figueroa JD, Kamat AM, Kiemeney L, Lotan Y, Pang K, Silverman DT et al: Epidemiology of Bladder Cancer: A Systematic Review and Contemporary Update of Risk Factors in 2018. Eur Urol 2018, 74(6): 784-795.
- Scélo G, Brennan P: The epidemiology of bladder and kidney cancer. Nat Clin Pract Urol 2007, 4(4): 205-217.
- Miyazaki J, Nishiyama H: Epidemiology of urothelial carcinoma. Int J Urol 2017, 24(10): 730-734.
- Gaddam SJ, Chesnut GT: Testicular Cancer. In: StatPearls. Epub ahead of print., edn. Treasure Island (FL): StatPearls Publishing Copyright © 2024, StatPearls Publishing LLC 2024.
- Ieda T, Muto S, Shimizu F, Taguri M, Yanada S, Kitamura K, Terai K, Saito K, Ogishima T, Nagata M et al: Development and Validation of a Novel Recurrence Risk Stratification for Initial Non-muscle Invasive Bladder Cancer in Asia. EBioMedicine 2016, 12: 98-104.
- Madeb R, Messing EM: Gender, racial and age differences in bladder cancer incidence and mortality. In: Urologic Oncology: Seminars and Original Investigations: 2004: Elsevier, 2004: 86-92.
- Kirkali Z, Chan T, Manoharan M, Algaba F, Busch C, Cheng L, Kiemeney L, Kriegmair M, Montironi R, Murphy WM et al: Bladder cancer: epidemiology, staging and grading, and diagnosis. Urology 2005, 66(6 Suppl 1): 4-34.
- Jalanko T, de Jong JJ, Gibb EA, Seiler R, Black PC: Genomic Subtyping in Bladder Cancer. Curr Urol Rep 2020, 21(2): 9.
- Al Aboud NM, Tupper C, Jialal I: Genetics, Epigenetic Mechanism. In: StatPearls. Epub ahead of print., edn. Treasure Island (FL): StatPearls Publishing Copyright © 2024, StatPearls Publishing LLC 2024.
- Pulumati A, Pulumati A, Dwarakanath BS, Verma A, Papineni RVL: Technological advancements in cancer diagnostics: Improvements and limitations. Cancer Rep (Hoboken) 2023, 6(2): e1764.
- Heyns CF, van der Merwe A: Bladder cancer in Africa. Can J Urol 2008, 15(1): 3899-3908.
- Penson DF: Socioeconomic factors, urological epidemiology and practice patterns. J Urol 2010, 184(5): 2094-2095.
- Koo KM, Mainwaring PN, Tomlins SA, Trau M: Merging new-age biomarkers and nanodiagnostics for precision prostate cancer management. Nat Rev Urol 2019, 16(5): 302-317.
- Wezel F, Bolenz CJRMCLC: Advances in medical treatments for genitourinary cancers. Revista Médica Clínica Las Condes 2018, 29(2): 155-162.
- Karemaker ID, Vermeulen M: Single-Cell DNA Methylation Profiling: Technologies and Biological Applications. Trends Biotechnol 2018, 36(9): 952-965.
- Gouil Q, Keniry A: Latest techniques to study DNA methylation. Essays Biochem 2019, 63(6): 639-648.
Annals of urologic oncology
p-ISSN: 2617-7765, e-ISSN: 2617-7773
 Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript