Research Article | Open Access
Long Non-coding RNA FAM66C Promotes Prostate Cancer Metastasis via JNK-Mediated Proteasome and Lysosomal Pathway
Zhenqian Qin1, Kongdong Li2, Jie Gu2, Yimin Xie1, Xuefeng Yuan1
1Department of Urology, Affiliated Hospital of Jiangsu University-Yixing Hospital, Yixing, Jiangsu, China.
2School of Life Sciences, Jiangsu University, Zhenjiang, Jiangsu, China.
Correspondence: Yimin Xie (Department of Urology, Affiliated Hospital of Jiangsu University-Yixing Hospital, Yixing, Jiangsu, 214200, China; Email: 2221513084@stmail.ujs.edu.cn) and Xuefeng Yuan (Department of Urology, Affiliated Hospital of Jiangsu University-Yixing Hospital, Yixing, Jiangsu, 214200, China; Email: staff983@yxph.com).
Annals of Urologic Oncology 2022; 5(1): 42-51. https://doi.org/10.32948/auo.2022.11.23
Received: 16 Nov 2022 | Accepted: 21 Nov 2022 | Published online: 29 Nov 2022
Methods The Cancer Genome Atlas (TCGA) data was utilized to determine the relative expression of lncRNA FAM66C in prostate cancer patients with lymph node metastasis. Knockdown FAM66C by siRNA was performed to investigate the effects of FAM66C in cell migration and epithelial-mesenchymal transition (EMT) by wound healing assay and Western blotting. The proteasome inhibitor MG132 and lysosomal inhibitor chloroquine (CQ) were used to determine the effect of these pathways in FAM66C-regulated cell migration. The c-jun-N-Terminal Kinase (JNK) inhibitor SP600125 was used to identify the role of JNK signaling in FAM66C-regulated cell migration and the proteasome and lysosome pathways.
Results A lower expression of lncRNA FAM66C was noted in the most prostate cancer patients. Knockdown of FAM66C in human prostate cancer DU145 and PC-3 cells promoted EMT and cell migration, which was suppressed by proteasomal inhibitor MG132 and lysosomal inhibitor CQ. Knockdown of FAM66C induced JNK signaling, cell migration and invasion, and activation of proteasome and lysosome pathways were suppressed by JNK inhibitor SP600125.
Conclusion This study provided new evidence of the role of lncRNA FAM66C in the regulation of JNK signaling mediated proteasome and lysosome pathways affecting migration ability of prostate cancer cells.
Key words LncRNA, prostate cancer, proteasome, Lysosome JNK, metastasis
Long non-coding RNA (lncRNA) is currently shown to have powerful potential as biomarkers for detection and therapeutic target(s) of cancers. It has reported that some lncRNAs promote prostate cancer metastasis, including lncRNA HOXD-AS1, Hottip, and PVT1 [3-5], and others such as lncRNA H19 suppress prostate cancer metastasis [6]. Most of these previous studies focused on identifying the oncogenic target(s) or miRNAs of these lncRNAs in cancers. However, the roles of these lncRNAs in cellular processes and underlying mechanisms in cancer progression are still unclear.
Lysosomal and proteasomal degradation are two major processes of protein degradation pathways that regulate degradation of metastasis proteins. These two pathways have been extensively studied and widely recognized as anticancer targets. Stable down-modulation of prosaposin decreases metastatic prostate cancer cell adhesion, migration, and invasion via a lysosomal proteolysis-dependent pathway [7]. Inhibition of cathepsin H, a lysosomal cysteine protease, reduced the processing of talin and migration of prostate cancer cells [8]. In addition, activation of retinoic acid receptor (RAR) inhibits prostate cancer metastasis by increasing the proteasomal degradation of the Smad1-RARγ complex, and thus blocking bone morphogenetic protein (BMP)-induced endothelial-to-osteoblast transition [9]. LncRNA SLC26A4-AS1 suppresses thyroid cancer metastasis by promoting DDX5 degradation through the ubiquitin-proteasome pathway [10]. Moreover, lncRNA LINRIS maintains the stability of a N6-methyladenosine (m6A) 'reader', insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2), and promotes the aerobic glycolysis in colorectal cancer by blocking ubiquitination and preventing the degradation of the autophagy-lysosome pathway [11]. However, the studies of the lysosomal and proteasomal degradations in lncRNA-regulated prostate cancer metastasis are still limited.
Our previous study showed that knockdown of lncRNA FAM66C increased ubiquitination and promote lysosomal acidic environment [12]. In the present study, we probed the prostate cancer dataset TCGA and analyzed through cBioPortal. Our analysis identified lower expression lncRNA FAM66C in prostate cancer patients with or without lymph node metastasis. By small interfering RNA transfection, we found that FAM66C knockdown significantly increased prostate cancer migration and EMT, and inactivated JNK signaling, which mediates the proteasomal ubiquitination and autophagy-lysosome pathways and promotes prostate cancer cell migration. The investigation of these two pathways involved in the lncRNAs regulated metastasis may be useful to understand the mechanism of prostate cancer progression.
Prostate cancer DU145 and PC-3 cell lines were obtained from American Tissue Culture Collection (ATCC, Manassas, VA, USA). DU145 and PC-3 cells were cultured in in RPMI 1640 (pH 7.4) medium, and supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, and 100 mg/ml streptomycin (pH 7.4) in a humidified atmosphere containing 5 % CO2. Mediums, FBS, penicillin, and streptomycin were purchased from Gibco (CA, USA). For some experiments, the cells were treated with JNK inhibitor, SP600125 (10 μM Tocris, UK), proteasomal inhibitor MG132 (10 μM, Sigma, USA), or lysosomal inhibitor Chloroquine (CQ, 20 μM, Sigma, USA) for 24 h.
Western blotting analysis
The cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (Beyotime, Shanghai, China). After centrifugation at 13, 000 g for 15 min at 4 0C, the supernatant was collected and the total protein concentration was determined by BCA (BCA Protein quantitative kit) (Thermo Fisher Scientific, USA). The protein lysates containing 30 μg total cellular protein in RIPA buffer were subjected to electrophoresis on 10% polyacrylamide gels. The gels were then blotted onto PVDF membranes (Merck, Shanghai, China). Western blotting was conducted using rabbit polyclonal antibodies against E-cadherin, N-cadherin, Vimentin, LC3B, P62 (1:1000, Cell Signaling Technology, Shanghai, China), anti-ubiquitin antibody (Santa Cruz, CA) and β-Actin (BM3883, Boster, Wuhan China), followed by incubation with horseradish peroxidase-conjugated goat anti-rabbit antibody (1:4000) (7074S, Cell Signaling Technology, Shanghai, China). Specific bands were visualized using chemiluminescent reagent (Vazyme, Nanjing, China).
Cell migration detection
Cell migratory capacity was assessed using the wound-healing assay. Cells were seeded in 6-wells plate (2 x 105 cells/well) in 2 mL complete medium. Once reaching 90-95% confluence, a scratch was created manually through the confluent monolayer using a white tip (2-20 μL). The culture medium was then aspirated, and wells were washed with PBS to remove cellular debris. Fresh medium without FBS was then added along with the indicated treatments. Images were taken at baseline (0 h) and at the indicated time points by Olympus IX73 microscopy (Japan). Microscope software was used to measure the width of the scratch, which reflected the extent of cell migration. The distance migrated was measured in μm.
Cell invasion assay
The cells were split and seeded at 2 × 105 cells in Matrigel-coated inserts with 8 μm pore size (BD Biosciences) of the up compartment. Cell invasion was evaluated after 24 h incubation. Non-invading cells on the upper side of the insert membrane were removed with cotton swabs. The invaded cells were fixed in ice-cold methanol for 10 min, and stained with 0.1% crystal violet for 20 min, then photographed and counted under microscope.
Knockdown of FAM66C
DU145 and PC-3 cells were seeded in 6-well with a density of 2 × 105 cells/well and cultured overnight. Then the cells were transfected with FAM66C siRNAs packaged by Lipo2000 reagent according to manufacturer instructions (Thermo Fisher Scientific, WI, USA). After 24 hrs transfection the cells were harvested for further evaluation. FAM66C siRNA: sense 5?-GCCUGCAAAGGUGAAUCAATT-3? and antisense 5?-UUGAUUCACCUUUGCAGGCTT-3?; and vector control: sense 5?-UUCUCCGAACGUGUCACGUTT-3?, antisense 5?-ACGUGACACGUUCGGAGAATT-3? from GenePharm (Shanghai, China).
Acridine orange (AO) staining
Acridine Orange (AO), a fluorochrome, emits red fluorescence in cell acidic compartments such as lysosomes or autophagolysosomes and green fluorescence in the cytosol and the nucleus. The ratio between the red and green signal of AO indicates the acidic characteristics of lysosomes. DU145 and PC-3 cells were seeded in 6-well and transfected with vector control, FAM66C siRNA-2 for 24 h, following treated with or without SP600125 for 24 h. After washed with PBS twice and AO (Leagene, Beijing, China) stain buffer once, the cells were stained by AO (1 mM) for 15 min at room temperature, and then fluorescence micrographs were obtained by Olympus BX53 (Japan). Quantification of the ratio between the red and green signal of AO was performed by using the software Image J.
Lysosomal acidity determination
LysoTrackerTM Green DND-26 is green fluorescent probe, which can indicate the acid environment in lysosome ( Thermo Fisher Scientific, USA). After treatment, Caki-1 cells washed with PBS twice, the cells were stained by LysoTrackerTM Green DND-26 (50 nM) in CO2 incubator for 15 min at 37 oC. Fluorescent intensity was recorded by excitation at 488 nm and emission at 525 nm using a Synergy H4 Hybrid Multi-Mode Microplate Reader (BioTek, USA).
Statistical analysis
Prostate cancer dataset (TCGA) (https://www.genenames. org/) was searched where 2708 lncRNAs (http://www.genenames.org) and 2687 lncRNAs (http://www. cbioportal.org) were analyzed by two different software. We selected all 499 tumors samples (498 patients) identified in the primary search. The primary search parameters included mutations, copy number alterations (CNA) from GISTIC and mRNA expression (RNA seq data) with the default setting. For secondary analysis, we focused on RNA seq data and analyzed the correlation between their expressions and the survival time of prostate cancer patients. All available lncRNAs were sorted by alteration frequency at the cBioPortal. Those lncRNAs with significant log-rank p values were entered in the candidate pool to be considered for further selection. P values less than 5% were determined as significant. Association with the FAM66C expression levels and clinic-pathological features were analyzed with the chi-square (χ2) test for categorical variables and the Wilcoxon test for continuous variables. The statistical analyses were performed using the SPSS 20.0 software analysis. The student t-test was used to compare the results and expressed as mean ± S.D. between control and treatment groups. P < 0.05 was considered statistically significant.
As reported in our previous study, FAM66C was identified as its expression correlated with patient survival times, tumor stage and Gleason pattern [12]. Moreover, it also showed that lower FAM66C expression associated with prostate cancer in patients with lymph node metastasis (N0, N1 or above) (Figure 1A). To detect the effects of FAM66C in prostate cancer migration, wound-healing assay was performed in FAM66C siRNA knockdown cells. The results showed that FAM66C knockdown resulted in significant promotion of migration of prostate cancer cells DU145 and PC-3 (Figure 1B-D). Western blotting analysis revealed that E-cadherin was reduced but N-cadherin was upregulated in FAM66C knockdown PC-3 cells (Figure 1E, F). Although no detectable N-cadherin was determined in DU145 cells, it showed decreased E-cadherin whereas Vimentin protein increased in FAM66C knockdown DU145 cells (Figure 1E, F).
Lower FAM66C expression promotes migration of prostate cancer cells through the proteasomal and lysosomal pathways
To identify the effects of FAM66C-regulated proteasomal and lysosomal pathways in migration of prostate cancer cells, wound-healing assay was performed in FAM66C siRNA knockdown cells with or without proteasomal inhibitor MG132 and lysosomal inhibitor CQ treatments. The results of showed that FAM66C knockdown-promoted migration of DU145 and PC-3 cells which was suppressed by MG132 and CQ (Figure 2A-D). These results indicate that lower FAM66C promotes migration of prostate cancer cells through the proteasomal and lysosomal pathways.
JNKK signaling mediates lower FAM66C-promoted cell migration
KEGG analysis of the co-expression genes of FAM66C indicated that JNK pathway might be involved in the migration of prostate cancer. Gene Expression Profiling Interactive Analysis (GEPIA) showed that FAM66C significantly positively correlated with JNK in prostate tumor (Figure 3A). Western blotting showed that siRNA knockdown FAM66C increased the protein level of p-JNK1/2 in DU145 cells (Figure 3B, C). In addition, results from the wound-healing experiments and transwell assay suggested that siRNA knockdown of FAM66C promoted cell migration and invasion that was significantly suppressed by JNK inhibitor SP600125 (Figure 3D-G). These results suggest that lower FAM66C expression promotes migration of prostate cancer cells depends on JNK activation.
FAM66C negatively regulates proteasome and lysosome pathways
To further clarify the roles of FAM66C-JNK on the proteasome and lysosome pathways, the proteasomal ubiquitin, autophagy biomarkers LC3B and P62 proteins levels were analyzed in FAM66C knockdown cells treated with or without JNK inhibitor SP600125. Western blotting showed that both the proteasome and lysosome pathways were inhibited by SP600125 (Figure 4A, B). In addition, we detected the acidic environment of lysosomes that is necessary for lysosomal degradation by Acridine Orange (AO) and lysosome tracker. Compared with control, the red fluorescence and the ratio of the red and green signal from AO increased in FAM66C knockdown cells, which was restored by SP600125 (Figure 4C, D). The lysosome tracker also showed that FAM66C siRNA increased lysosomal pH was reversed by SP600125 (Figure 4E, F). Taken together, these results suggest that JNK signaling mediates lower FAM66C-activated proteasomal and lysosomal pathways.
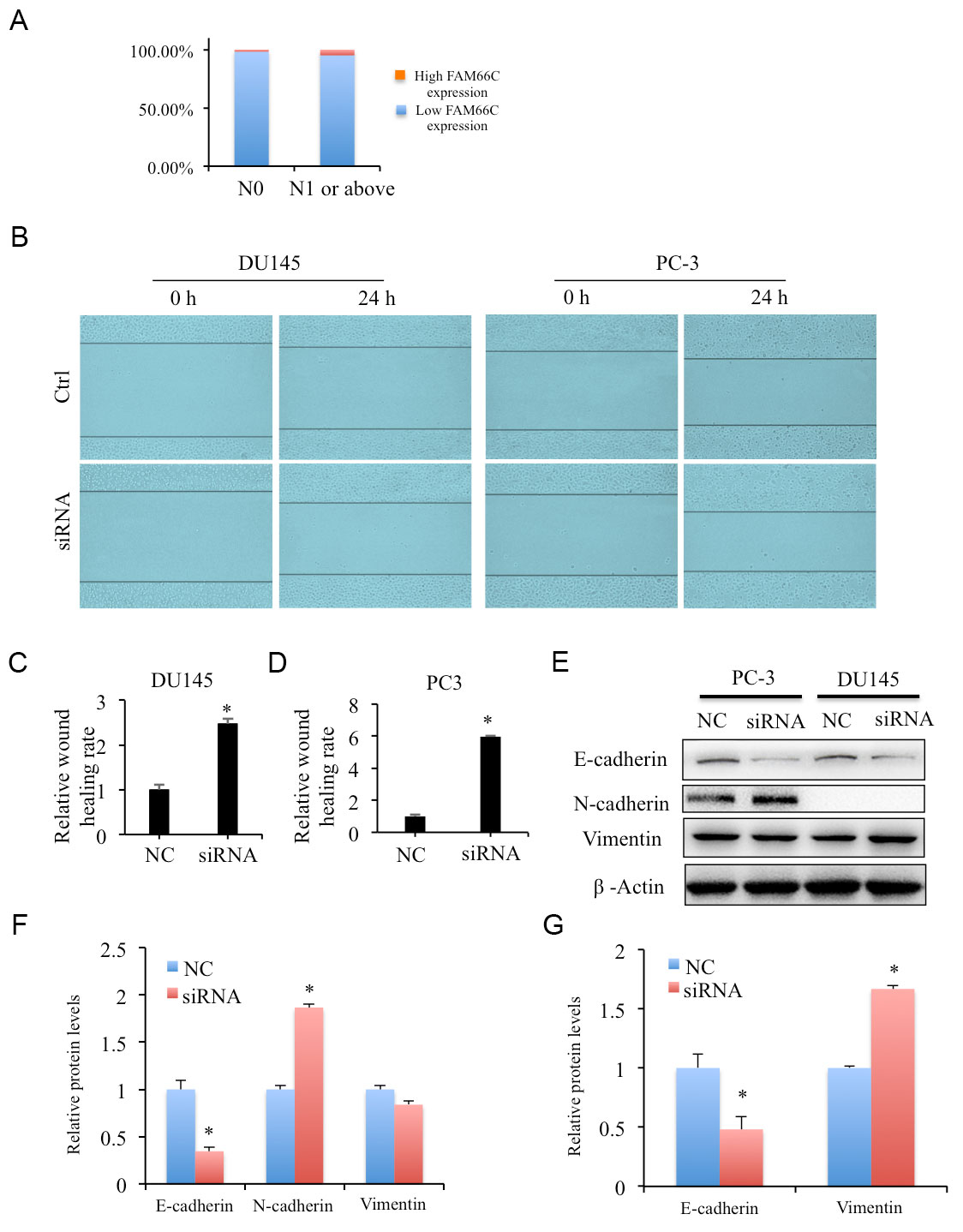 Figure 1. Lower expression of FAM66C promotes cell migration of prostate cancer cells. (A) Lower FAM66C expression associated with in prostate cancer patients with lymph node metastasis. (B-D) Effects of FAM66C knockdown by siRNA in DU145 and PC-3 cell migration detected by wound healing assays. (E, F) The protein levels of EMT biomarkers were determined by western blotting in FAM66C knockdown DU145 and PC-3 cells. β-Actin was used as loading reference. Quantification was performed by the software Image J. * P <0.05.
Figure 1. Lower expression of FAM66C promotes cell migration of prostate cancer cells. (A) Lower FAM66C expression associated with in prostate cancer patients with lymph node metastasis. (B-D) Effects of FAM66C knockdown by siRNA in DU145 and PC-3 cell migration detected by wound healing assays. (E, F) The protein levels of EMT biomarkers were determined by western blotting in FAM66C knockdown DU145 and PC-3 cells. β-Actin was used as loading reference. Quantification was performed by the software Image J. * P <0.05.
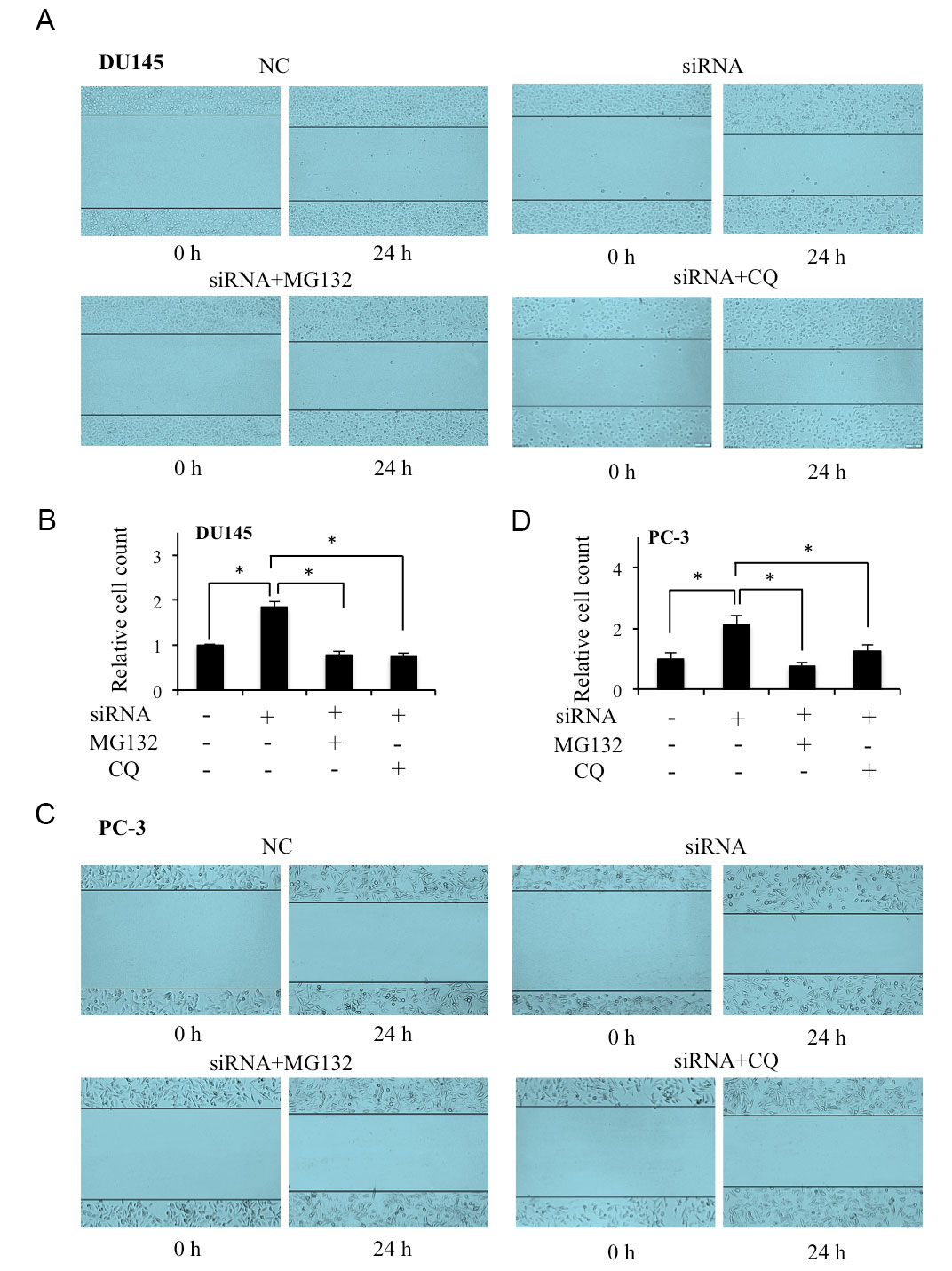 Figure 2. Effects of MG132 and CQ on lower FAM66C promoted migration of prostate cancer cells. (A-D) The cell migration were evaluated by wound healing assay after siRNA knockdown DU145 and PC-3 cells incubated with in the absence or presence of MG132, CQ for 24 h. Quantifications were analyzed by Image J. * P <0.05. MG132: proteasomal inhibitor, 10 μM, CQ: lysosomal inhibitor, 20 μM.
Figure 2. Effects of MG132 and CQ on lower FAM66C promoted migration of prostate cancer cells. (A-D) The cell migration were evaluated by wound healing assay after siRNA knockdown DU145 and PC-3 cells incubated with in the absence or presence of MG132, CQ for 24 h. Quantifications were analyzed by Image J. * P <0.05. MG132: proteasomal inhibitor, 10 μM, CQ: lysosomal inhibitor, 20 μM.
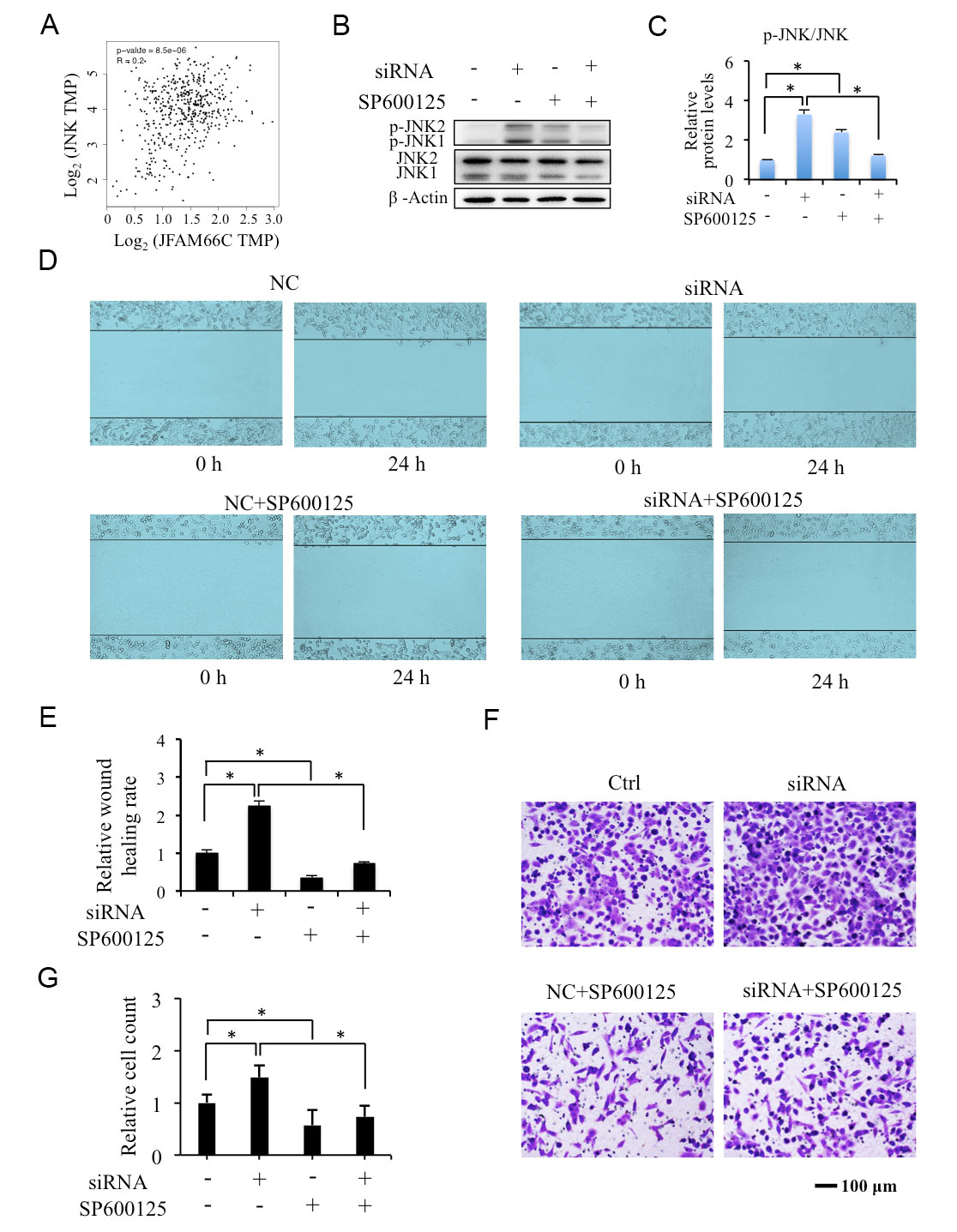 Figure 3. Effects of SP100625 on lower FAM66C promoted migration of prostate cancer cells. (A, B) Effects of FAM66C knockdown on p-JNK protein level in DU145 cells. β-Actin was used as loading reference. (C-F) The cell migration and invasion in FAM66C knockdown DU145 cells incubated with in the absence or presence of 10 μM SP600125 for 24 h detected by wound healing and transwell assays. Quantification was performed by the software Image J. * P <0.05.
Figure 3. Effects of SP100625 on lower FAM66C promoted migration of prostate cancer cells. (A, B) Effects of FAM66C knockdown on p-JNK protein level in DU145 cells. β-Actin was used as loading reference. (C-F) The cell migration and invasion in FAM66C knockdown DU145 cells incubated with in the absence or presence of 10 μM SP600125 for 24 h detected by wound healing and transwell assays. Quantification was performed by the software Image J. * P <0.05.
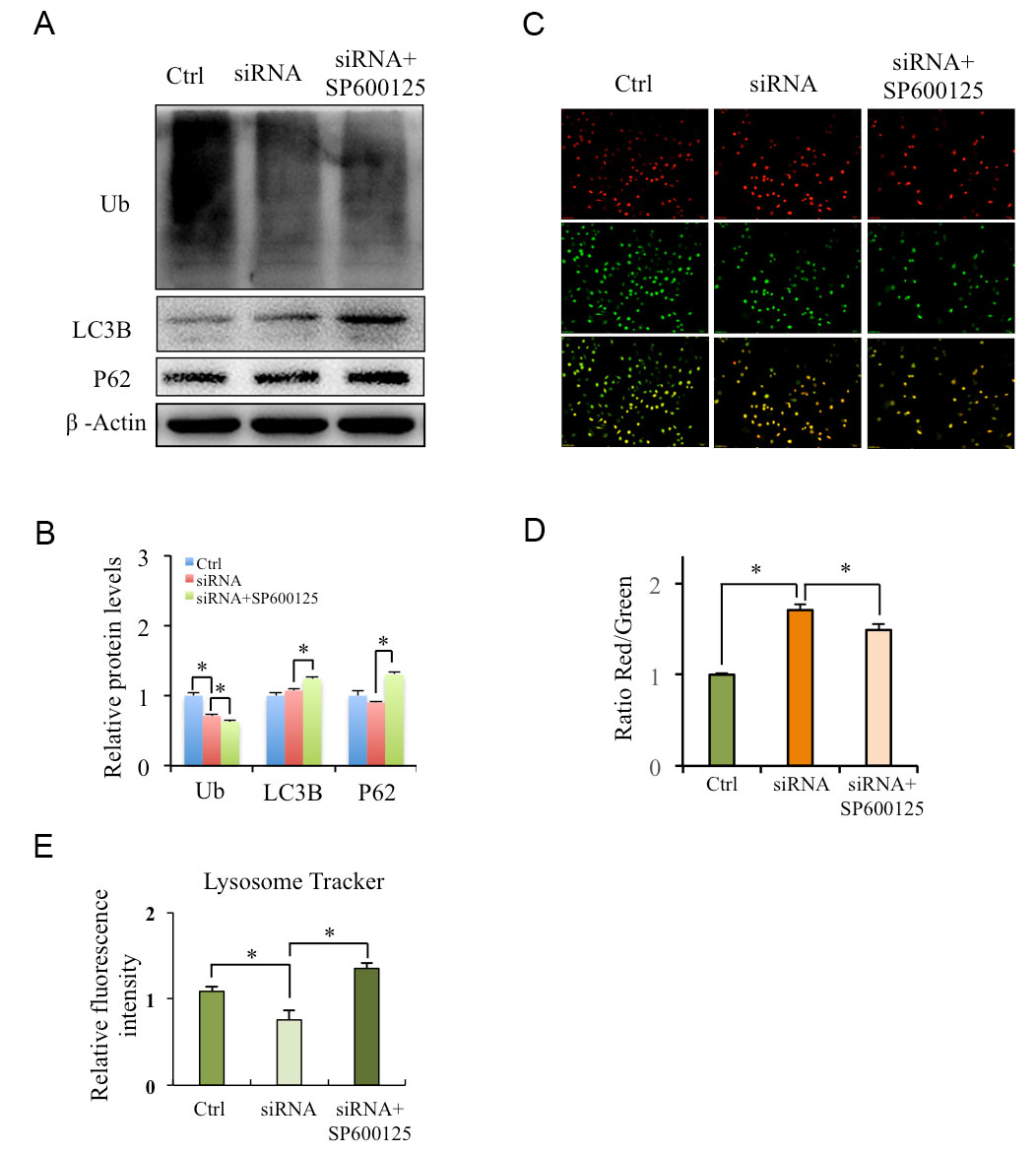 Figure 4. Roles of SP100625 on lower on FAM66C-modulated ubiquitination and lysosome acidity in prostate cancer cells. (A, B) Roles of SP100625 on the protein levels of ubiquitin and autophagy. FAM66C knockdown DU145 cells were treated with SP600125 for 24 h, the expressions of ubiquitin were detected by Western blotting. Quantification of relative protein levels was performed by the software Image J. (C, D) Roles of SP100625 on lysosomal acidity in FAM66C knockdown DU145 cells. FAM66C knockdown DU145cells were stained by AO, acidity region of lysosome appear Red. Quantification of the ratio between the red and green signal of AO was performed by the software Image J. (E) Lysosomal Tracker indicates the lysosome pH. * P < 0.05, using Student’s t-test.
Figure 4. Roles of SP100625 on lower on FAM66C-modulated ubiquitination and lysosome acidity in prostate cancer cells. (A, B) Roles of SP100625 on the protein levels of ubiquitin and autophagy. FAM66C knockdown DU145 cells were treated with SP600125 for 24 h, the expressions of ubiquitin were detected by Western blotting. Quantification of relative protein levels was performed by the software Image J. (C, D) Roles of SP100625 on lysosomal acidity in FAM66C knockdown DU145 cells. FAM66C knockdown DU145cells were stained by AO, acidity region of lysosome appear Red. Quantification of the ratio between the red and green signal of AO was performed by the software Image J. (E) Lysosomal Tracker indicates the lysosome pH. * P < 0.05, using Student’s t-test.
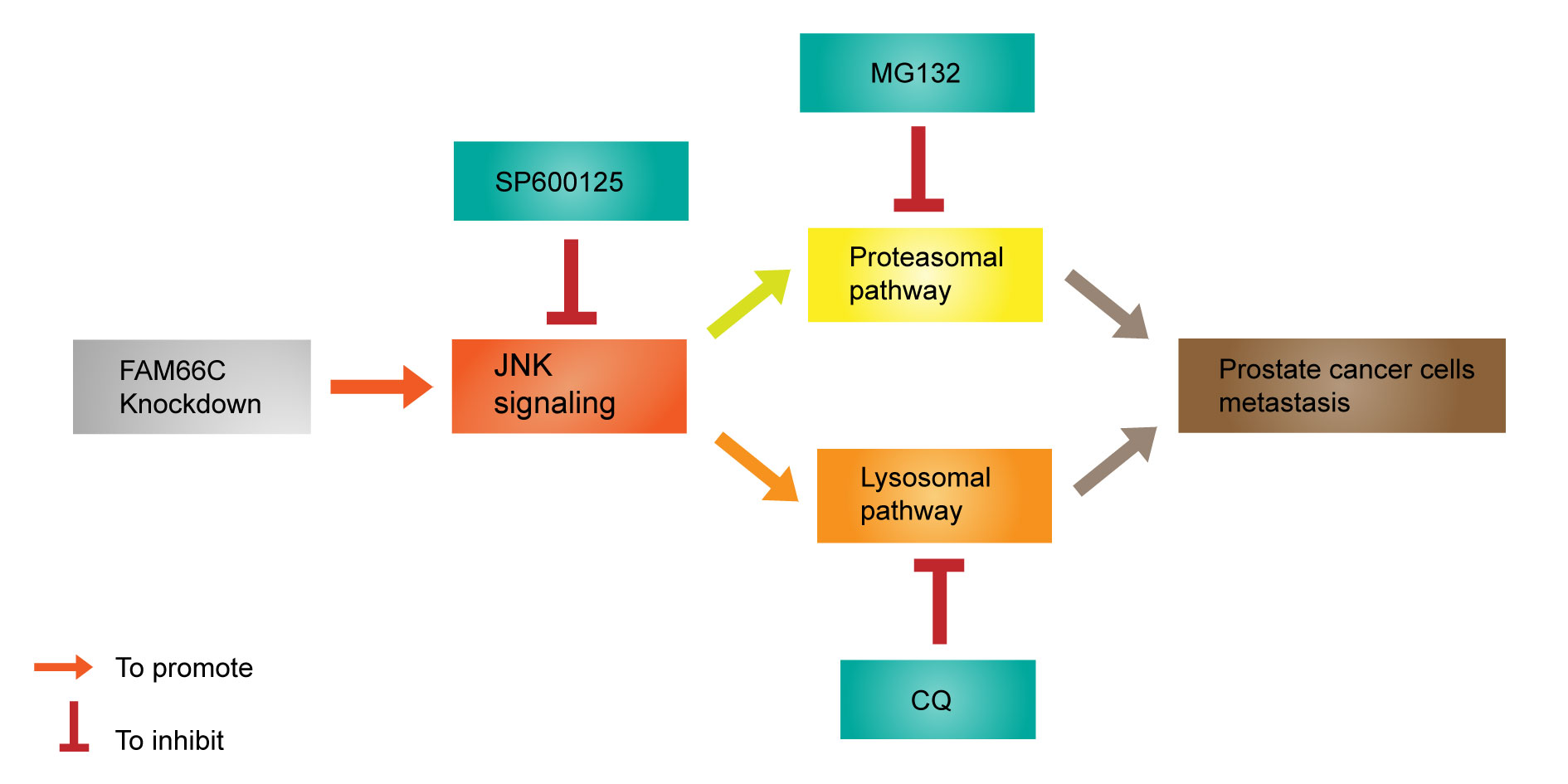 Figure 5. Schematic diagram to show the mechanism that relationship between LncRNA FAM66C and prostate cancer.
Figure 5. Schematic diagram to show the mechanism that relationship between LncRNA FAM66C and prostate cancer.
lncRNAs have been reported to play important roles in regulating cancer cell fate, such as growth, apoptosis, autophagy and metastasis [16]. The lncRNA expression level, such as LINC01296 was higher in prostate cancer tissues and prostate cancer cells than in the normal tissues or cells, and LINC01296 knockdown inhibited prostate cancer cell migration, invasion and EMT [17]. These lower lncRNAs expression is predictor of poor prognosis in prostate cancer patients. For example, the expression levels of lncRNAs HCG11 were significantly lower in prostate cancer tissues than those in non-tumor tissues, and associated with the lymph node status [18]. In addition, an integrative analysis revealed that downregulation of lncRNA PCAT-14 expression significantly associated with poor prognosis of prostate cancer patients. Low PCAT-14 expression increased cell migration, which was reduced in overexpressing PCAT-14 prostate cancer cells [19]. These studies suggest that both higher and lower lncRNA expression may promote cancer cell migration. Our results show that knockdown FAM66C promotes migration and EMT of prostate cancer cells. Interestingly, Sun et al. identified that F-box only protein 22 (FBXO22) was upregulated in breast tumors and promoted tumor growth but suppressed EMT and metastasis, suggesting FBXO22 played a dual role in mammary tumorigenesis and metastasis [20]. Taken together our previous study [12] and the present work show that lncRNA FAM66C might play a dual role in prostate tumor growth and metastasis.
Previous studies have shown that lncRNAs regulate the lysosomal and proteasomal pathways, which control the stability of metastatic proteins thus regulating cell migration. Therefore, lncRNAs might modulate migration of the cancer cells through the lysosomal and proteasomal pathways. LncRNA ANCR showed lower expression in breast cancer tissues and breast cancer cell lines, and knockdown of ANCR promoted EMT and cell migration and invasion, via inhibiting proteasomal degradation of EMT inducer EZH2 [21]. LncRNA DRAIC is downregulated in gastric cancer tissues and cell lines, which inhibits metastasis of gastric cancer cells via interfering with deubiquitination of NFRKB by UCHL5 [22]. lncRNA SLC26A4-AS1 was significantly downregulated not only in normal tissues but also in thyroid cancer, as well as in lymph node metastasis specimens. LncRNA SLC26A4-AS1 suppresses thyroid cancer metastasis by promoting DDX5 degradation through the ubiquitin-proteasome pathway [10]. On the other hand, lncRNA LINRIS was upregulated in colorectal cancer tissues and promotes the aerobic glycolysis by blocking ubiquitination and preventing the degradation of the autophagy-lysosome pathway [11]. LncRNA HAGLR and TP73-AS1 were up-regulated and promote EMT and metastasis by targeting increase lysosome-associated membrane glycoprotein 3 (LAMP3) and lysosomal-associated protein transmembrane-4 Beta (LAPTM4B) in esophageal cancer and lung cancer [23, 24]. LncRNA TOB1-AS1 was decreased in gastric cancer tissues and cell lines, which inhibited cell migration and invasion by co-regulating lysosomal sialidase NEU1 [25]. In the present study, the FAM66C knockdown promoted prostate cancer migration was abrogated by proteasomal inhibitor MG132 and lysosomal inhibitor CQ, which suggested that the lysosomal and proteasomal pathways mediated FAM66C-regulated prostate cancer migration.
In prostate cancer, the JNK pathway play significant roles in regulating various cell processes, including apoptosis, proliferation, migration and invasion, and JNK could be potential therapeutic target in prostate cancer [26]. It showed that neferine inhibited migration of human prostate cancer stem cells through JNK activation [27], and isoliquiritigenin inhibited migration and invasion of prostate cancer cells by decreasing JNK signaling activation [28]. It had reported that overexpression of a circular RNA circ-102004 caused cell migration and invasion of prostate cancer cells by upregulation of JNK and p-JNK [29]. In addition, the roles of lncRNAs-regulated JNK on the migration of the cancer cells have also been investigated. LINC00958 accelerated cell migration in non-small cell lung cancer through JNK signaling [30], and LINC00116 enhanced migration of cervical cancer cells through JNK pathway [31], while LINC01111 suppresses cell invasion and migration of pancreatic cancer by inactivating the JNK signaling pathway [32]. However, the regulation of lncRNA on JNK signaling in prostate cancer progression has not been previously identified. Our results showed that knockdown of FAM66C caused activation of JNK, which further mediated migration and invasion of prostate cancer cells.
Recent studies suggested that JNK positively regulates autophagy to counteract apoptosis and develop the chemotherapeutic resistance of cancer cells [33]. A complete autophagic flux or not refers to the lysosomal degradation or dysregulation, and JNK acts as a keeper of lysosomal stability in cancer cells [34, 35]. In triple-negative breast cancer, JNK Inhibitor JNK-IN-8 suppressed tumor growth by inducing lysosome biogenesis and autophagy [36]. Moreover, the proteasomal and autophagy-lysosomal degradation are two complementary pathways, and could be mediated by JNK. In lung cancer cells H1299, JNK-mediated the degradation of histone deacetylase 8 expression autophagy and the proteasome system [37]. The proteasome inhibitors suppressed the ubiquitin-proteasome system but activated autophagy by inducing activation of the JNK pathway in cancer cells [38]. In addition, the proteasome inhibitor MG132 induced activation of JNK signaling, and the JNK inhibitor SP600125 enhanced proteasome inhibitor MG132-induced apoptosis of kidney cancer cells [39]. In hepatocellular carcinoma cells, JNK inhibitor SP600125 specifically reduced the stability of the lysosomal membrane protein LAMP2A, which promoted its degradation by the proteasome and caused cell death [35]. These studies support that JNK might be the linker of lysosome and proteasome pathways. In the present study, knockdown of FAM66C-activated ubiquitin-proteasome and autophagy-lysosome pathways were significantly suppressed by JNK inhibitor.
None.
Ethical policy
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study. Approval from institutional ethical committee was taken.
Author contributions
ZQ, and KL performed the research; ZQ, JG, YX and XY designed the research study and contributed essential reagents or tools; ZQ, KL, JG, YX and XY analyzed and interpreted the data; ZQ, JG, YX and XY wrote the manuscript. All authors reviewed the manuscript.
Competing interests
All authors declare no competing interests.
Funding
This work was supported by the Foundation of Health and Family Planning Commission of Wuxi City (Q201807), the Foundation of Clinical Medical Science and Technology Development project of Jiangsu University (JLY20180005), the Foundation of Science and Technology Development Medical and Health Guidance Plan Project of Wuxi City (2020061), the Innovation Foundation of Science and Technology Bureau of Yixing City (2020SF08), the Municipal Health Commission Suitable Technology promotion project of Wuxi City (T202126), and the Municipal Health Commission Suitable Technology promotion project of Jiangsu province (LSD2022008).
- Xia C, Dong X, Li H, Cao M, Sun D, He S, Yang F, Yan X, Zhang S, Li N, et al: Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl) 2022, 135(5): 584-590.
- Altschuler J, Stockert JA, Kyprianou N: Non-Coding RNAs set a new phenotypic frontier in prostate cancer metastasis and resistance. Int J Mol Sci 2021, 22(4): 2100.
- Malek R, Gajula RP, Williams RD, Nghiem B, Simons BW, Nugent K, Wang H, Taparra K, Lemtiri-Chlieh G, Yoon AR, et al: TWIST1-WDR5-Hottip regulates hoxa9 chromatin to facilitate prostate cancer metastasis. Cancer Res 2017, 77(12): 3181-3193.
- Sun F, Wu K, Yao Z, Mu X, Zheng Z, Sun M, Wang Y, Liu Z, Zhu Y: Long noncoding RNA PVT1 promotes prostate cancer metastasis by increasing NOP2 expression via targeting tumor suppressor MicroRNAs. Onco Targets Ther 2020, 13: 6755-6765.
- Jiang Y, Zhao H, Chen Y, Li K, Li T, Chen J, Zhang B, Guo C, Qing L, Shen J, et al: Exosomal long noncoding RNA HOXD-AS1 promotes prostate cancer metastasis via miR-361-5p/FOXM1 axis. Cell Death Dis 2021, 12(12): 1129.
- Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J, et al: lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J 2014, 281(16): 3766-3775.
- Hu S, Delorme N, Liu Z, Liu T, Velasco-Gonzalez C, Garai J, Pullikuth A, Koochekpour S: Prosaposin down-modulation decreases metastatic prostate cancer cell adhesion, migration, and invasion. Mol Cancer 2010, 9: 30.
- Jevnikar Z, Rojnik M, Jamnik P, Doljak B, Fonovic UP, Kos J: Cathepsin H mediates the processing of talin and regulates migration of prostate cancer cells. J Biol Chem 2013, 288(4): 2201-2209.
- Yu G, Corn PG, Shen P, Song JH, Lee YC, Lin SC, Pan J, Agarwal SK, Panaretakis T, Pacifici M, et al: Retinoic acid receptor activation reduces metastatic prostate cancer bone lesions by blocking the endothelial-to-osteoblast transition. Cancer Res 2022, 82(17): 3158-3171.
- Yuan J, Song Y, Pan W, Li Y, Xu Y, Xie M, Shen Y, Zhang N, Liu J, Hua H, et al: LncRNA SLC26A4-AS1 suppresses the MRN complex-mediated DNA repair signaling and thyroid cancer metastasis by destabilizing DDX5. Oncogene 2020, 39(43): 6664-6676.
- Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen YX, Liu J, Luo XJ, Meng Q, Pu HY, et al: LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer 2019, 18(1): 174.
- 12. Xie Y, Gu J, Qin Z, Ren Z, Wang Y, Shi H, Chen B: Long non-coding RNA FAM66C is associated with clinical progression and promotes cell proliferation by inhibiting proteasome pathway in prostate cancer. Cell Biochem Funct 2020, 38(8): 1006-1016.
- Zhu J, Zhu S, Yu Q, Wu Y: LncRNA FAM66C inhibits pancreatic cancer progression by sponging miR-574-3p. Transl Cancer Res 2020, 9(3): 1806-1817.
- Lei GL, Li Z, Li YY, Hong ZX, Wang S, Bai ZF, Sun F, Yan J, Yu LX, Yang PH, et al: Long noncoding RNA FAM66C promotes tumor progression and glycolysis in intrahepatic cholangiocarcinoma by regulating hsa-miR-23b-3p/KCND2 axis. Environ Toxicol 2021, 36(11): 2322-2332.
- Xiao K, Peng G: Long non-coding RNA FAM66C regulates glioma growth via the miRNA/LATS1 signaling pathway. Biol Chem 2022, 403(7): 679-689.
- Yang G, Lu X, Yuan L: LncRNA: a link between RNA and cancer. Biochim Biophys Acta 2014, 1839(11): 1097-1109.
- Wu J, Cheng G, Zhang C, Zheng Y, Xu H, Yang H, Hua L: Long noncoding RNA LINC01296 is associated with poor prognosis in prostate cancer and promotes cancer-cell proliferation and metastasis. Onco Targets Ther 2017, 10: 1843-1852.
- Zhang Y, Zhang P, Wan X, Su X, Kong Z, Zhai Q, Xiang X, Li L, Li Y: Downregulation of long non-coding RNA HCG11 predicts a poor prognosis in prostate cancer. Biomed Pharmacother 2016, 83: 936-941.
- White NM, Zhao SG, Zhang J, Rozycki EB, Dang HX, McFadden SD, Eteleeb AM, Alshalalfa M, Vergara IA, Erho N, et al: Multi-institutional Analysis Shows that Low PCAT-14 Expression Associates with Poor Outcomes in Prostate Cancer. Eur Urol 2017, 71(2): 257-266.
- Sun R, Xie HY, Qian JX, Huang YN, Yang F, Zhang FL, Shao ZM, Li DQ: FBXO22 possesses both protumorigenic and antimetastatic roles in breast cancer progression. Cancer Res 2018, 78(18): 5274-5286.
- Li Z, Hou P, Fan D, Dong M, Ma M, Li H, Yao R, Li Y, Wang G, Geng P, et al: The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ 2017, 24(1): 59-71.
- Zhang Z, Hu X, Kuang J, Liao J, Yuan Q: LncRNA DRAIC inhibits proliferation and metastasis of gastric cancer cells through interfering with NFRKB deubiquitination mediated by UCHL5. Cell Mol Biol Lett 2020, 25: 29.
- Jiang Q, Xing W, Cheng J, Yu Y: Long Non-Coding RNA TP73-AS1 Promotes the Development of Lung Cancer by Targeting the miR-27b-3p/LAPTM4B Axis. Onco Targets Ther 2020, 13: 7019-7031.
- Yang C, Shen S, Zheng X, Ye K, Sun Y, Lu Y, Ge H: Long noncoding RNA HAGLR acts as a microRNA-143-5p sponge to regulate epithelial-mesenchymal transition and metastatic potential in esophageal cancer by regulating LAMP3. FASEB J 2019, 33(9): 10490-10504.
- Jiang K, Zhi XH, Ma YY, Zhou LQ: Long non-coding RNA TOB1-AS1 modulates cell proliferation, apoptosis, migration and invasion through miR-23a/NEU1 axis via Wnt/b-catenin pathway in gastric cancer. Eur Rev Med Pharmacol Sci 2019, 23(22): 9890-9899.
- Xu R, Hu J: The role of JNK in prostate cancer progression and therapeutic strategies. Biomed Pharmacother 2020, 121: 109679.
- Erdogan S, Turkekul K: Neferine inhibits proliferation and migration of human prostate cancer stem cells through p38 MAPK/JNK activation. J Food Biochem 2020, 44(7): e13253.
- Kwon GT, Cho HJ, Chung WY, Park KK, Moon A, Park JH: Isoliquiritigenin inhibits migration and invasion of prostate cancer cells: possible mediation by decreased JNK/AP-1 signaling. J Nutr Biochem 2009, 20(9): 663-676.
- Si-Tu J, Cai Y, Feng T, Yang D, Yuan S, Yang X, He S, Li Z, Wang Y, Tang Y, et al: Upregulated circular RNA circ-102004 that promotes cell proliferation in prostate cancer. Int J Biol Macromol 2019, 122: 1235-1243.
- Luo Z, Han Z, Shou F, Li Y, Chen Y: LINC00958 accelerates cell proliferation and migration in non-small cell lung cancer through JNK/c-JUN signaling. Hum Gene Ther Methods 2019, 30(6): 226-234.
- Lai YQ, Zhou BB, Tan QQ, Xu J, Wan T, Zhang L: LINC00116 enhances cervical cancer tumorigenesis through miR-106a/c-Jun pathway. J Cell Biochem 2020, 121(3): 2247-2257.
- Pan S, Shen M, Zhou M, Shi X, He R, Yin T, Wang M, Guo X, Qin R: Long noncoding RNA LINC01111 suppresses pancreatic cancer aggressiveness by regulating DUSP1 expression via microRNA-3924. Cell Death Dis 2019, 10(12): 883.
- Wu Q, Wu W, Fu B, Shi L, Wang X, Kuca K: JNK signaling in cancer cell survival. Med Res Rev 2019, 39(6): 2082-2104.
- Zhou YY, Li Y, Jiang WQ, Zhou LF: MAPK/JNK signalling: a potential autophagy regulation pathway. Biosci Rep 2015, 35(3): e00199.
- Desideri E, Ciriolo MR: Inhibition of JNK increases the sensitivity of hepatocellular carcinoma cells to lysosomotropic drugs via LAMP2A destabilization. Cell Death Discov 2021, 7(1): 29.
- Soleimani M, Somma A, Kaoud T, Goyal R, Bustamante J, Wylie DC, Holay N, Looney A, Giri U, Triplett T, et al: Covalent JNK inhibitor, JNK-IN-8, suppresses tumor growth in triple-negative breast cancer by activating TFEB- and TFE3-mediated lysosome biogenesis and autophagy. Mol Cancer Ther 2022, 21(10): 1547-1560.
- Park JY, Juhnn YS: cAMP signaling increases histone deacetylase 8 expression by inhibiting JNK-dependent degradation via autophagy and the proteasome system in H1299 lung cancer cells. Biochem Biophys Res Commun 2016, 470(2): 336-342.
- Wu WK, Sakamoto KM, Milani M, Aldana-Masankgay G, Fan D, Wu K, Lee CW, Cho CH, Yu J, Sung JJ: Macroautophagy modulates cellular response to proteasome inhibitors in cancer therapy. Drug Resist Updat 2010, 13(3): 87-92.
- Wang HQ, Liu BQ, Gao YY, Meng X, Guan Y, Zhang HY, Du ZX: Inhibition of the JNK signalling pathway enhances proteasome inhibitor-induced apoptosis of kidney cancer cells by suppression of BAG3 expression. Br J Pharmacol 2009, 158(5): 1405-1412.
Annals of urologic oncology
p-ISSN: 2617-7765, e-ISSN: 2617-7773
 Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript