Review Article | Open Access
Research Progress on Circulating Tumor DNA in Renal Cell Carcinoma
Xinyi Zhang1, Mohan Dong2, Kaiyuan Zhou3
1Quality Control Department, Shaanxi Provincial Hospital of Traditional Chinese Medicine, Xi’an, China.
2Medical Department, the First Affiliated Hospital, Air Force Medical University, Xi’an, China.
3Health Service Department, Air Force Medical University, Xi’an, China.
Correspondence: Kaiyuan Zhou (Health Service Department, Air Force Medical University, 169 Changle West Street, Xi’an, 710032, China; Email: csnrzky@126.com) and Mohan Dong (Medical Department, the First Affiliated Hospital, Air Force Medical University, 127 Changle West Street, Xi’an, 710032, China; Email: 715674449@qq.com).
Annals of Urologic Oncology 2023, 6(2): 54-62. https://doi.org/10.32948/auo.2023.06.10
Received: 30 May 2023 | Accepted: 10 Jun 2023 | Published online: 16 Jun 2023
Key words Circulating tumor DNA, gene mutation, methylation, renal cell carcinoma, clinical research
Cell-free DNA (cfDNA) refers to nucleation that can be detected in almost all body fluids, including blood.The short DNA fragments are involved in various physiological and pathological processes such as immunity, coagulation, aging, carcinogenesis, etc. In cancer patients, a portion of the cfDNA in the plasma originates from the tumor, called circulating tumor DNA (ctDNA) , and may have the same mutations and genetic alterations as the primary tumor. However, ctDNA detection technique provides a new opportunity to solve the above problem. ctDNA is a fragment of DNA in the blood or body fluids derived from tumor cells, it is mainly derived from apoptosis, necrosis and extracellular vesicle secretion of tumor cells [18-34]. It is a characteristic tumor biomarker. By analyzing the characteristics of ctDNA, a wide range of information about the tumor can be obtained. The core advantage of ctDNA detection is that it reflects the information of the entire tumor genome [35, 36], and it is real-time and easy to obtain. This technique avoids clinical biopsy puncture damage and the internal heterogeneity of the tumor, the tumor molecular typing is more accurate. The genomic information obtained from patient's guide the course of comprehensive treatment may provide a basis for adjusting clinical treatment plans. Furthermore, patients can be monitored noninvasively to detect prospective tumor progression. Thus, the application value of ctDNA in the fight against cancer has received great attention in recent years, but the application of ctDNA in the clinical diagnosis and treatment of RCC is still in the early stage of exploration. Further study of its clinical value will benefit patients at all stages of RCC and help to provide personalized treatment for mRCC. This review introduces the clinical research progress of ctDNA in the diagnosis of RCC, prognosis assessment, disease recurrence monitoring, therapeutic effect monitoring and acquired drug resistance.
|
Table 1. Tumor-guided analysis of plasma. |
||||
|
Study |
Stage |
Methods |
ctDNA detection rate (%) |
Findings |
|
Maia MC 2017 [38] |
mRCC |
CLIA-certified, College of American Pathology-accredited comprehensive plasma assay; The enriched digital sequence libraries were analyzed using the HiSeq2500 Sequencing System (Illumina) with an average coverage depth of 15,000 x. |
53 |
The content of ctDNA in peripheral blood of mRCC patients is correlated with tumor load, and the tumor volume of these patients is significantly larger than that of patients without ctDNA detected. |
|
Wan J 2013 [39] |
Various |
Quantitative real-time PCR was performed on an ABI Prism ® 7900HT. Each 10-μl reaction consisted of 1XSYBR ® GreenER TM quantitative PCR SuperMix, 1 μl DNA sample, and 0.2 μmol/l forward and reverse primers (ACTB: forward GCTATCCCTGTACGCCTCTG; reverse AGGAAGGAAGGCTGGAAGAG; size of PCR product is 384 bp). |
NA |
The level of plasma ctDNA is associated with the recurrence of RCC, and the recurrence rate of tumor in patients with high plasma ctDNA level before nephrectomy is significantly higher than that in patients with low plasma ctDNA level. |
|
Feng G 2013[40] |
mRCC |
Quantitative real-time PCR was performed on an ABI Prism 7900 HT. |
NA |
Plasma ctDNA levels were negatively correlated with prognosis during treatment. |
|
Yamamoto Y 2018 [41] |
various |
Quantitative real-time PCR analysis was performed using a CFX Connect™ Real-Time System (Bio-Rad Laboratories, Hercules, CA, USA) to detect levels of plasma cfDNA. |
63 |
The median plasma ctDNA level in RCC patients was significantly higher than that in healthy control group, and the plasma ctDNA level increased with the increase of tumor TNM stage and Fuhrman grade. The ctDNA level in patients with lymphatic vessel infiltration was also significantly higher than that in patients without invasion. Compared with healthy controls, plasma ctDNA levels are significantly higher even in patients with early (cT1aN0M0) RCC. |
|
Yamamoto 2019 [42] |
Various |
Target capture and further library preparation pro -cesses were carried out according to the manufacturer's instructions for the Agilent SureSelectXT Target Enrichment System (Agilent Technologies) with minor modification. |
30 |
Plasma ctDNA fragment length was significantly shorter in RCC patients than in healthy controls, and ctDNA fragment size was inversely associated with progression-free survival in RCC patients, with shorter mean ctDNA length in patients with higher Fuhrman grade and positive lymphatic infiltration. |
|
Khagi Y 2017 [44] |
Various |
Sequencing was performed by a Clinical Laboratory Improvement Amendments (CLIA)-certified and College of American Pathologists (CAP)-accredited clinical laboratory, Guardant Health, Inc. (http://www.guardanthealth.com). |
91 |
ctDNA is derived from genomic DNA, and the number of base mutations carried by ctDNA is directly related to the mutation load of a patient's tumor cells. Detecting the mutation profile of CTDNA can help predict whether a patient can benefit from immunological checkpoint inhibitor therapy. |
|
Pal SK 2017 2018 [43, 50] |
Various |
Enriched digital sequence libraries are subsequently analyzed using the HiSeq2500 Sequencing System (Illumina, San Diego, CA, USA), achieving an average raw coverage depth of 15 000 (minimum: 2000, average Q-score: 20). |
78.6 |
The ctDNA mutant profile of 220 patients with mRCC and classified it to assess whether there were significant differences in the reactivity of patients with different ctDNA mutations to targeted therapy.the increased mutation level of p53 gene in ctDNA was associated with resistance to Sunitinib, pazopanib, bevacizumab and other targeted drugs. |
|
Lu H 2016 [57] |
Various |
DNA was quantified using Quant-iT™ PicoGreen® dsDNA Reagent (ThermoFisher, Invitrogen, Darmstadt, Germany) in Greiner 384 well plates (Sigma-Aldrich, Taufkirchen, Germany) on the microplate reader Mithras LB 940 (Berthold Technologies, Bad Wildbad, Germany). |
NA |
The plasma cfDNA and cfmtDNA concentrations of 145 non-metastatic RCC patients and 84 mRCC patients, and established a diagnostic model of RCC by combining the concentration of nuclear genome and mitochondrial genome (AUC up to 0.84). |
|
Skrypkina I 2016 [26] |
Various |
The methylation status of the different genes was determined qualitatively by the methylation-specific polymerase chain reaction (MS-PCR). |
LRRC3B (74.1%), APC (51.9%), FHIT (55.6%), and RASSF1 (62.9%) genes |
Ras association domain family 1A could be detected in the plasma DNA of RCC patients (ras association domain family 1A, RASSF1A), fragile histidine triad (FHIT) and adenomatous polyposis coli (APC) methylation having great value in the diagnosis of RCC. |
|
Jung M 2019 [58] |
Various |
Quantification of SHOX2 DNA methylation in ccfDNA and tissues from the UKB cohort was conducted using a methylation-specific quantitative PCR (qMSP). |
NA |
The methylation level of short stature homeobox protein 2 (SHOX2) in RCC tissue and plasma samples and evaluated its clinical significance, It was found that patients with RCC who are still at high risk of death after nephrectomy can be identified by testing SHOX2 methylation levels in ctDNA before treatment, and may benefit from adjuvant therapy or early palliative care through early intervention. |
|
Costa VL 2011 [60] |
Various |
Chemiluminescence detection, image acquisition and analysis were performed using an Applied Biosystems Chemiluminescence Detection Kit (P/N 4342142) and Applied Biosystems 1700 Chemiluminescent Microarray Analyzer (P/N 4338036) following the manufacturer’s protocol. |
67 |
PCDH17 methylation in urine samples from RCC patients has the potential to be a biomarker for urinary tumors, including bladder cancer, RCC, and prostate cancer. |
|
Xin J 2016 [61] |
Various |
Following PCR amplification, pyrosequencing was performed using a PyroMark Gold Q96 SQA Reagents kit (Qiagen, Inc.) and a PSQ96 HS DNA analyzing system (Qiagen, Inc.) according to the manufacturer's protocol. |
89 |
The level of TCF21 in the urine of RCC patients positively correlate with its level in tumor tissue, and has certain diagnostic value for RCC. |
|
Santoni M 2018 [62] |
Various |
The genomic DNA of patients was extracted from blood, using the DNA Blood Mini kit according to the manufacturer’s instructions. Genotyping of polymorphisms was performed using pre-designed TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s instructions. Amplifications and analysis were carried out on the 7300 Real-Time PCR System (Applied Biosystems), using the SDS software v1.4.0 for allelic discrimination (Applied Biosystems). |
NA |
The genotype of autophagy genes in peripheral blood to evaluate its correlation with the risk and prognosis of renal clear cell carcinoma. |
|
NA: Not Applicable. |
||||
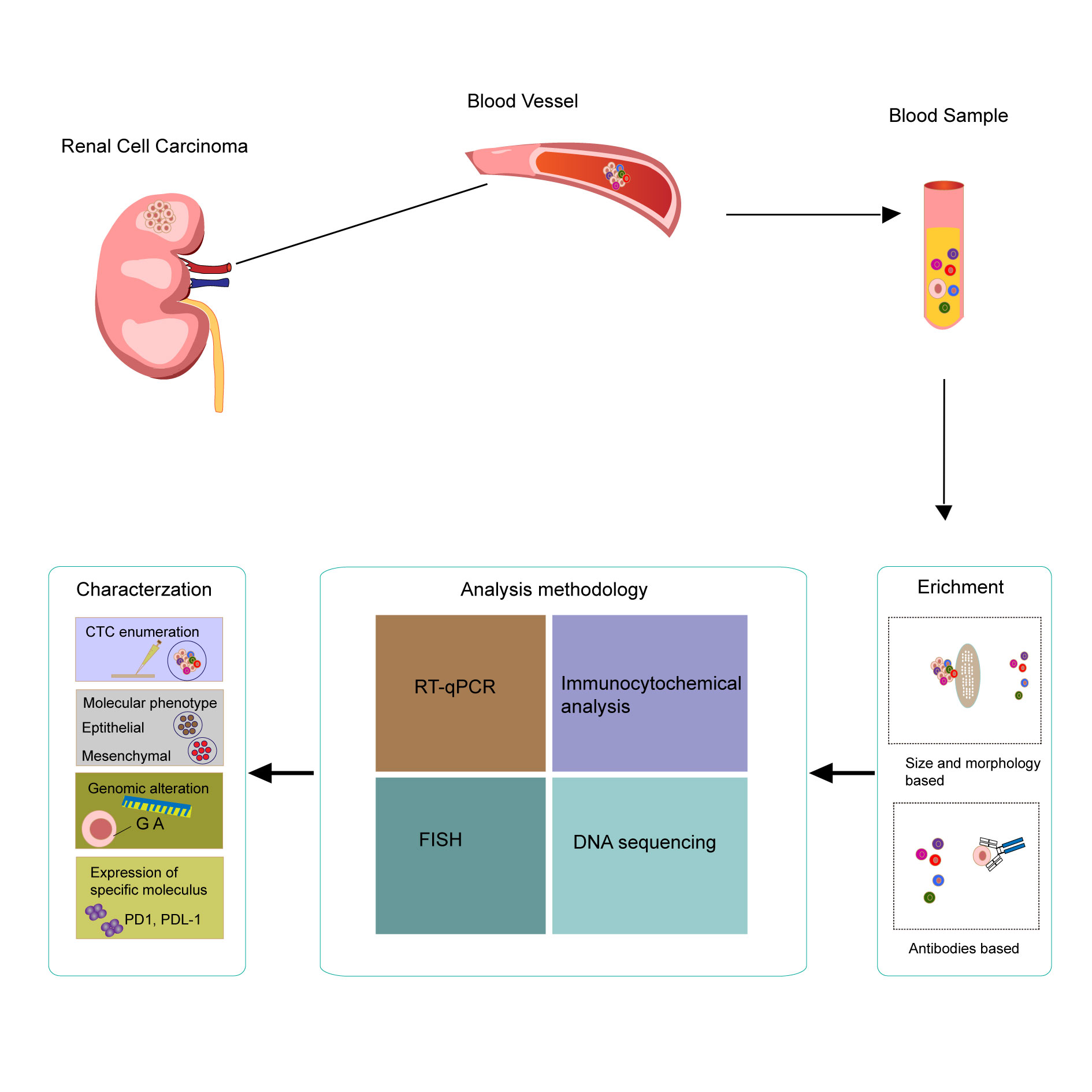 Figure 1. Samples for liquid biopsy of RCC using bood samples comprising circulating tumor DNA (ctDNA), and isolation, detection, and characterization of ctDNA and the associated clinical value.
Figure 1. Samples for liquid biopsy of RCC using bood samples comprising circulating tumor DNA (ctDNA), and isolation, detection, and characterization of ctDNA and the associated clinical value.
Autophagy plays an important regulatory role in the occurrence and progression of RCC. Santoni et al. [62] analyzed the genotype of autophagy genes in peripheral blood to evaluate its correlation with the risk and prognosis of renal clear cell carcinoma. The autophagyrelated genes selected included ATG4A, ATG4B, ATG4C, ATG5, ATG16L1, ATG16L2 and IRGM in autophagyrelated gene (ATG). Single nucleotide polymorphism (SNP) of these genes was analyzed in 40 mRCC patients treated with pazopanib, and the incidence of ATG16L2-rs10751215 was significantly reduced in mRCC patients compared with the general population. ATG4A-rs7880351, ATG4Crs6670694, and ATG5-rs490010 were associated with disease-progression-free survival in pazopanib treated RCC patients.
Mitochondria
Lu et al. [57] investigated the application potential of genomic and mitochondrial derived ctDNA fragments in the diagnosis and prognosis of RCC patients. They extracted ctDNA from 40 healthy controls and 229 patients with RCC (145 RCC and 84 mRCC) for analysis. Two mitochondrial fragments (65 bp and 175 bp) were found to be able to effectively distinguish healthy controls from patients with RCC and mRCC. And one genomic ctDNA fragment (306 bp) was found to be able to distinguish between healthy controls and patients with RCC. These results suggest that genomic and mitochondrial ctDNA fragments may contribute to the early diagnosis of mRCC, which may have important applications in adjuvant therapy of RCC.
We thank Dr. Sanjay Gupta (Case Western Reserve University & UH Cleveland Medical Center) for his proofreading for the review.
Ethical policy
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study. Approval from institutional ethical committee was taken.
Availability of data and materials
All data generated or analysed during this study are included in this publication.
Author contributions
XZ wrote the manuscript draft and prepared the figures and tables. MD and KZ edited and approved the final manuscript.
Competing interests
All authors declare no competing interests.
Funding
None.
- Siegel RL, Miller KD, Jemal A: Cancer statistics, 2018. CA Cancer J Clin 2018, 68(1): 7-30.
- Janzen NK, Kim HL, Figlin RA, Belldegrun AS: Surveillance after radical or partial nephrectomy for localized renal cell carcinoma and management of recurrent disease. Urol Clin North Am 2003, 30(4): 843-852.
- Escudier B, Porta C, Schmidinger M, Rioux-Leclercq N, Bex A, Khoo V, Gruenvald V, Horwich A: Renal cell carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016, 27(suppl 5): v58-v68.
- Schwaab T, Schwarzer A, Wolf B, Crocenzi TS, Seigne JD, Crosby NA, Cole BF, Fisher JL, Uhlenhake JC, Mellinger D et al: Clinical and immunologic effects of intranodal autologous tumor lysate-dendritic cell vaccine with Aldesleukin (Interleukin 2) and IFN-{alpha}2a therapy in metastatic renal cell carcinoma patients. Clin Cancer Res 2009, 15(15): 4986-4992.
- De Meerleer G, Khoo V, Escudier B, Joniau S, Bossi A, Ost P, Briganti A, Fonteyne V, Van Vulpen M, Lumen N et al: Radiotherapy for renal-cell carcinoma. Lancet Oncol 2014, 15(4): e170-177.
- Motzer RJ, Bander NH, Nanus DM: Renal-cell carcinoma. N Engl J Med 1996, 335(12): 865-875.
- Singh P, Agarwal N, Pal SK: Sequencing systemic therapies for metastatic kidney cancer. Curr Treat Options Oncol 2015, 16(1): 316.
- Kroog GS, Motzer RJ: Systemic therapy for metastatic renal cell carcinoma. Urol Clin North Am 2008, 35(4): 687-701.
- de Velasco G, Hamieh L, Mickey S, Choueiri TK: Optimizing systemic therapy for metastatic renal cell carcinoma beyond the first-line setting. Urol Oncol 2015, 33(12): 538-545.
- Choueiri TK, Motzer RJ: Systemic Therapy for Metastatic Renal-Cell Carcinoma. N Engl J Med 2017, 376(4):354-366.
- Larroquette M, Peyraud F, Domblides C, Lefort F, Bernhard JC, Ravaud A, Gross-Goupil M: Adjuvant therapy in renal cell carcinoma: Current knowledges and future perspectives. Cancer Treat Rev 2021, 97: 102207.
- Mitsogiannis IC, Mitsogianni M, Papathanassiou M, Anagnostou M, Tamposis I, Mitrakas L, Samara M, Tzortzis V, Vlachostergios PJ: Current Options for Second-Line Systemic Therapy in Metastatic Renal Cell Carcinoma. J Kidney Cancer VHL 2022, 9(3): 29-40.
- Logan JE, Rampersaud EN, Sonn GA, Chamie K, Belldegrun AS, Pantuck AJ, Slamon DJ, Kabbinavar FF: Systemic therapy for metastatic renal cell carcinoma: a review and update. Rev Urol 2012, 14(3-4): 65-78.
- Cavaliere C, D'Aniello C, Pepa CD, Pisconti S, Berretta M, Facchini G: Current and Emerging Treatments for Metastatic Renal Cell Carcinoma. Curr Cancer Drug Targets 2018, 18(5): 468-479.
- Motzer RJ, Tannir NM, McDermott DF, Arén Frontera O, Melichar B, Choueiri TK, Plimack ER, Barthélémy P, Porta C, George S et al: Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. N Engl J Med 2018, 378(14): 1277-1290.
- Powles T, Lackner MR, Oudard S, Escudier B, Ralph C, Brown JE, Hawkins RE, Castellano D, Rini BI, Staehler MD et al: Randomized Open-Label Phase II Trial of Apitolisib (GDC-0980), a Novel Inhibitor of the PI3K/Mammalian Target of Rapamycin Pathway, Versus Everolimus in Patients With Metastatic Renal Cell Carcinoma. J Clin Oncol 2016, 34(14): 1660-1668.
- Shin SJ, Kim T, Sung HH, Jeon HG, Jeong BC, Park SH, Jeon SS, Lee HM, Choi HY, Seo SI et al: Novel Predictive Models of Early Death Less Than 1 Year in Patients With Metastatic Renal Cell Carcinoma After Treatment With First-line Tyrosine Kinase Inhibitors. Clin Genitourin Cancer 2019, 17(6): e1137-e1146.
- Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M: Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev 2016, 35(3): 347-376.
- Jiang P, Chan CW, Chan KC, Cheng SH, Wong J, Wong VW, Wong GL, Chan SL, Mok TS, Chan HL et al: Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A 2015, 112(11): E1317-1325.
- An Q, Hu Y, Li Q, Chen X, Huang J, Pellegrini M, Zhou XJ, Rettig M, Fan G: The size of cell-free mitochondrial DNA in blood is inversely correlated with tumor burden in cancer patients. Precis Clin Med 2019, 2(3): 131-139.
- Kim YJ, Kang Y, Kim JS, Sung HH, Jeon HG, Jeong BC, Seo SI, Jeon SS, Lee HM, Park D et al: Potential of circulating tumor DNA as a predictor of therapeutic responses to immune checkpoint blockades in metastatic renal cell carcinoma. Sci Rep 2021, 11(1): 5600.
- Bacon JVW, Annala M, Soleimani M, Lavoie JM, So A, Gleave ME, Fazli L, Wang G, Chi KN, Kollmannsberger CK et al: Plasma Circulating Tumor DNA and Clonal Hematopoiesis in Metastatic Renal Cell Carcinoma. Clin Genitourin Cancer 2020, 18(4): 322-331.
- Zengin ZB, Weipert C, Salgia NJ, Dizman N, Hsu J, Meza L, Chehrazi-Raffle A, Muddasani R, Salgia S, Malhotra J et al: Complementary Role of Circulating Tumor DNA Assessment and Tissue Genomic Profiling in Metastatic Renal Cell Carcinoma. Clin Cancer Res 2021, 27(17): 4807-4813.
- Hahn AW, Gill DM, Maughan B, Agarwal A, Arjyal L, Gupta S, Streeter J, Bailey E, Pal SK, Agarwal N: Correlation of genomic alterations assessed by next-generation sequencing (NGS) of tumor tissue DNA and circulating tumor DNA (ctDNA) in metastatic renal cell carcinoma (mRCC): potential clinical implications. Oncotarget 2017, 8(20): 33614-33620.
- Hauser S, Zahalka T, Fechner G, Müller SC, Ellinger J: Serum DNA hypermethylation in patients with kidney cancer: results of a prospective study. Anticancer Res 2013, 33(10): 4651-4656.
- Skrypkina I, Tsyba L, Onyshchenko K, Morderer D, Kashparova O, Nikolaienko O, Panasenko G, Vozianov S, Romanenko A, Rynditch A: Concentration and Methylation of Cell-Free DNA from Blood Plasma as Diagnostic Markers of Renal Cancer. Dis Markers 2016, 2016: 3693096.
- Hauser S, Zahalka T, Ellinger J, Fechner G, Heukamp LC, A VONR, Müller SC, Bastian PJ: Cell-free circulating DNA: Diagnostic value in patients with renal cell cancer. Anticancer Res 2010, 30(7): 2785-2789.
- Gang F, Guorong L, An Z, Anne GP, Christian G, Jacques T: Prediction of clear cell renal cell carcinoma by integrity of cell-free DNA in serum. Urology 2010, 75(2): 262-265.
- Heitzer E, Haque IS, Roberts CES, Speicher MR: Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet 2019, 20(2): 71-88.
- Corcoran RB, Chabner BA: Application of Cell-free DNA Analysis to Cancer Treatment. N Engl J Med 2018, 379(18): 1754-1765.
- Cimadamore A, Massari F, Santoni M, Mollica V, Di Nunno V, Cheng L, Lopez-Beltran A, Scarpelli M, Montironi R, Moch H: Molecular characterization and diagnostic criteria of renal cell carcinoma with emphasis on liquid biopsies. Expert Rev Mol Diagn 2020, 20(2): 141-150.
- Chin RI, Chen K, Usmani A, Chua C, Harris PK, Binkley MS, Azad TD, Dudley JC, Chaudhuri AA: Detection of Solid Tumor Molecular Residual Disease (MRD) Using Circulating Tumor DNA (ctDNA). Mol Diagn Ther 2019, 23(3): 311-331.
- Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA et al: Circulating mutant DNA to assess tumor dynamics. Nat Med 2008, 14(9): 985-990.
- To EW, Chan KC, Leung SF, Chan LY, To KF, Chan AT, Johnson PJ, Lo YM: Rapid clearance of plasma Epstein-Barr virus DNA after surgical treatment of nasopharyngeal carcinoma. Clin Cancer Res 2003, 9(9): 3254-3259.
- Li G, Pavlick D, Chung JH, Bauer T, Tan BA, Peguero J, Ward P, Kallab A, Bufill J, Hoffman A et al: Genomic profiling of cell-free circulating tumor DNA in patients with colorectal cancer and its fidelity to the genomics of the tumor biopsy. J Gastrointest Oncol 2019, 10(5): 831-840.
- Li H, Di Y, Li J, Jiang Y, He H, Yao L, Gu J, Lu J, Song J, Chen S et al: Blood-based Genomic Profiling of Circulating Tumor DNA from Patients with Advanced Pancreatic Cancer and its Value to Guide Clinical Treatment. J Cancer 2020, 11(15): 4316-4323.
- Diaz LA, Bardelli A: Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014, 32(6): 579-586.
- Maia MC, Bergerot PG, Dizman N, Hsu J, Jones J, Lanman RB, Banks KC, Pal SK: Association of Circulating Tumor DNA (ctDNA) Detection in Metastatic Renal Cell Carcinoma (mRCC) with Tumor Burden. Kidney Cancer 2017, 1(1): 65-70.
- Wan J, Zhu L, Jiang Z, Cheng K: Monitoring of plasma cell-free DNA in predicting postoperative recurrence of clear cell renal cell carcinoma. Urol Int 2013, 91(3): 273-278.
- Feng G, Ye X, Fang F, Pu C, Huang H, Li G: Quantification of plasma cell-free DNA in predicting therapeutic efficacy of sorafenib on metastatic clear cell renal cell carcinoma. Dis Markers 2013, 34(2): 105-111.
- Yamamoto Y, Uemura M, Nakano K, Hayashi Y, Wang C, Ishizuya Y, Kinouchi T, Hayashi T, Matsuzaki K, Jingushi K et al: Increased level and fragmentation of plasma circulating cell-free DNA are diagnostic and prognostic markers for renal cell carcinoma. Oncotarget 2018, 9(29): 20467-20475.
- Yamamoto Y, Uemura M, Fujita M, Maejima K, Koh Y, Matsushita M, Nakano K, Hayashi Y, Wang C, Ishizuya Y et al: Clinical significance of the mutational landscape and fragmentation of circulating tumor DNA in renal cell carcinoma. Cancer Sci 2019, 110(2): 617-628.
- Pal SK, Sonpavde G, Agarwal N, Vogelzang NJ, Srinivas S, Haas NB, Signoretti S, McGregor BA, Jones J, Lanman RB et al: Evolution of Circulating Tumor DNA Profile from First-line to Subsequent Therapy in Metastatic Renal Cell Carcinoma. Eur Urol 2017, 72(4): 557-564.
- Khagi Y, Goodman AM, Daniels GA, Patel SP, Sacco AG, Randall JM, Bazhenova LA, Kurzrock R: Hypermutated Circulating Tumor DNA: Correlation with Response to Checkpoint Inhibitor-Based Immunotherapy. Clin Cancer Res 2017, 23(19): 5729-5736.
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS et al: Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357(6349): 409-413.
- Choueiri TK, Hessel C, Halabi S, Sanford B, Michaelson MD, Hahn O, Walsh M, Olencki T, Picus J, Small EJ et al: Cabozantinib versus sunitinib as initial therapy for metastatic renal cell carcinoma of intermediate or poor risk (Alliance A031203 CABOSUN randomised trial): Progression-free survival by independent review and overall survival update. Eur J Cancer 2018, 94: 115-125.
- Gill D, Hahn AW, Sonpavde G, Agarwal N: Immunotherapy of advanced renal cell carcinoma: Current and future therapies. Hum Vaccin Immunother 2016, 12(12): 2997-3004.
- Zhao H, Nolley R, Chan AMW, Rankin EB, Peehl DM: Cabozantinib inhibits tumor growth and metastasis of a patient-derived xenograft model of papillary renal cell carcinoma with MET mutation. Cancer Biol Ther 2017, 18(11): 863-871.
- Beuselinck B, Jean-Baptiste J, Schöffski P, Couchy G, Meiller C, Rolland F, Allory Y, Joniau S, Verkarre V, Elaidi R et al: Validation of VEGFR1 rs9582036 as predictive biomarker in metastatic clear-cell renal cell carcinoma patients treated with sunitinib. BJU Int 2016, 118(6): 890-901.
- Pal SK, Ali SM, Yakirevich E, Geynisman DM, Karam JA, Elvin JA, Frampton GM, Huang X, Lin DI, Rosenzweig M et al: Characterization of Clinical Cases of Advanced Papillary Renal Cell Carcinoma via Comprehensive Genomic Profiling. Eur Urol 2018, 73(1): 71-78.
- Brodziak A, Sobczuk P, Bartnik E, Fiedorowicz M, Porta C, Szczylik C, Czarnecka AM: Drug resistance in papillary RCC: from putative mechanisms to clinical practicalities. Nat Rev Urol 2019, 16(11): 655-673.
- Zhou J, Luo J, Wu K, Yun EJ, Kapur P, Pong RC, Du Y, Wang B, Authement C, Hernandez E et al: Loss of DAB2IP in RCC cells enhances their growth and resistance to mTOR-targeted therapies. Oncogene 2016, 35(35): 4663-4674.
- Schröck A, Leisse A, de Vos L, Gevensleben H, Dröge F, Franzen A, Wachendörfer M, Schröck F, Ellinger J, Teschke M et al: Free-Circulating Methylated DNA in Blood for Diagnosis, Staging, Prognosis, and Monitoring of Head and Neck Squamous Cell Carcinoma Patients: An Observational Prospective Cohort Study. Clin Chem 2017, 63(7): 1288-1296.
- Tham C, Chew M, Soong R, Lim J, Ang M, Tang C, Zhao Y, Ong SY, Liu Y: Postoperative serum methylation levels of TAC1 and SEPT9 are independent predictors of recurrence and survival of patients with colorectal cancer. Cancer 2014, 120(20): 3131-3141.
- Fenner A: Kidney cancer: Methylation panel predicts RCC outcome. Nat Rev Urol 2017, 14(1): 8-9.
- Wei JH, Haddad A, Wu KJ, Zhao HW, Kapur P, Zhang ZL, Zhao LY, Chen ZH, Zhou YY, Zhou JC et al: A CpG-methylation-based assay to predict survival in clear cell renal cell carcinoma. Nat Commun 2015, 6: 8699.
- Lu H, Busch J, Jung M, Rabenhorst S, Ralla B, Kilic E, Mergemeier S, Budach N, Fendler A, Jung K: Diagnostic and prognostic potential of circulating cell-free genomic and mitochondrial DNA fragments in clear cell renal cell carcinoma patients. Clin Chim Acta 2016, 452: 109-119.
- Jung M, Ellinger J, Gevensleben H, Syring I, Lüders C, de Vos L, Pützer S, Bootz F, Landsberg J, Kristiansen G et al: Cell-Free SHOX2 DNA Methylation in Blood as a Molecular Staging Parameter for Risk Stratification in Renal Cell Carcinoma Patients: A Prospective Observational Cohort Study. Clin Chem 2019, 65(4): 559-568.
- Lin YL, Wang YP, Li HZ, Zhang X: Aberrant Promoter Methylation of PCDH17 (Protocadherin 17) in Serum and its Clinical Significance in Renal Cell Carcinoma. Med Sci Monit 2017, 23: 3318-3323.
- Costa VL, Henrique R, Danielsen SA, Eknaes M, Patrício P, Morais A, Oliveira J, Lothe RA, Teixeira MR, Lind GE et al: TCF21 and PCDH17 methylation: An innovative panel of biomarkers for a simultaneous detection of urological cancers. Epigenetics 2011, 6(9): 1120-1130.
- Xin J, Xu R, Lin S, Xin M, Cai W, Zhou J, Fu C, Zhen G, Lai J, Li Y et al: Clinical potential of TCF21 methylation in the diagnosis of renal cell carcinoma. Oncol Lett 2016, 12(2): 1265-1270.
- Santoni M, Piva F, De Giorgi U, Mosca A, Basso U, Santini D, Buti S, Lolli C, Terrone C, Maruzzo M et al: Autophagic Gene Polymorphisms in Liquid Biopsies and Outcome of Patients with Metastatic Clear Cell Renal Cell Carcinoma. Anticancer Res 2018, 38(10): 5773-5782.
- Ball MW, Gorin MA, Guner G, Pierorazio PM, Netto G, Paller CJ, Hammers HJ, Diaz LA, Allaf ME: Circulating Tumor DNA as a Marker of Therapeutic Response in Patients With Renal Cell Carcinoma: A Pilot Study. Clin Genitourin Cancer 2016, 14(5): e515-e520.
- Corrò C, Hejhal T, Poyet C, Sulser T, Hermanns T, Winder T, Prager G, Wild PJ, Frew I, Moch H et al: Detecting circulating tumor DNA in renal cancer: An open challenge. Exp Mol Pathol 2017, 102(2): 255-261.
Annals of urologic oncology
p-ISSN: 2617-7765, e-ISSN: 2617-7773
 Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Ann Urol Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript